Abstract
Striated muscle has a highly ordered structure in which specialized domains of the cell membrane involved in force transmission (costameres) and excitation-contraction coupling (T tubules) as well as the internal membranes of the sarcoplasmic reticulum are organized over specific regions of the sarcomere. Optimal muscle function is dependent on this high level of organization but how it established and maintained is not well understood. Due to its ex utero development and transparency, the zebrafish embryo is an excellent vertebrate model for the study of dynamic relationships both within and between cells during development. Transgenic models have allowed the delineation of cellular migration and complex morphogenic rearrangements during the differentiation of skeletal myocytes and the assembly and organization of new myofibrils. Molecular targeting of genes and transcripts has allowed the identification of key requirements for myofibril assembly and organization. With the recent advances in gene editing approaches, the zebrafish will become an increasingly important model for the study of human myopathies and muscular dystrophies. Its high fecundity and small size make it well suited to high-throughput screenings to identify novel pharmacologic and molecular therapies for the treatment of a broad range of neuromuscular conditions. In this review, we examine the lessons learned from the zebrafish model regarding the complex interactions between the sarcomere and the sarcolemma that pattern the developing myocyte and discuss the potential for zebrafish as a model system to examine the pathophysiology of, and identify new treatments for, human myopathies and muscular dystrophies.
Keywords: myofibrils, T tubules, sarcoplasmic reticulum, sarcolemma, zebrafish, skeletal muscle
Myofibril organization
The highly ordered structure of vertebrate striated muscle is required for its normal function. Precise relationships between the contractile structures of the myofibril and the surrounding cytoskeletal and membrane compartments are maintained during contraction and relaxation. The cytoskeletal framework that surrounds and supports the myofibril links the myofibrils to each other and to specialized regions of the cell membrane and the sarcoplasmic reticulum involved in force transmission and excitation-contraction coupling, respectively (Hein et al., 2000). The importance of maintaining this structure is highlighted by the number of myopathies and muscular dystrophies that result when these relationships are altered (Kanagawa and Toda, 2006; Capetanaki et al., 2007; Kontrogianni-Konstantopoulos et al., 2009). Yet how this highly organized structure is established is not well understood.
What has been determined is that there is a close relationship between newly assembling myofibrils and the sarcolemma. Non-striated fibrils, representing immature or pre-myofibrils, were first noted immediately adjacent to the cell membrane in trout skeletal muscle [reviewed in Sanger et al. (2010)]. This finding has been confirmed in skeletal myotubes, in remodeling cardiac myocytes in vitro, in chick cardiac explants in primary culture, and in the developing human heart suggesting that interactions between the membrane and the myofibril promote the early steps of striated myofibril assembly in both heart and skeletal muscle (Terai et al., 1989; Kim et al., 1992; Rhee et al., 1994). The close association between the myofibril and the membrane [and through transmembrane adhesive contacts, the extracellular matrix (ECM)] may promote coordination of their maturation and organization during development and during adaptive remodeling. However, it has been difficult to model the development of the complex cellular organization of striated myocyte in vitro due to the importance of the three dimensional structure of surrounding cells and tissues in establishing the cell's internal architecture. In vivo studies have yielded important insights into cell and tissue organization but higher vertebrate models have been limited by the difficulty with monitoring the process in the developing embryo.
The zebrafish as a model system for skeletal muscle development
The zebrafish (Danio rerio) is an excellent in vivo model system in which to examine the dynamic processes of new myofibril assembly and myocyte organization during development. As a vertebrate, the sarcomeric and cytoskeletal proteins involved in these processes are highly conserved with those in higher vertebrates including humans. Furthermore, the dynamics and progression of myofibril assembly has been demonstrated to be highly similar to that observed in mammalian models (Sanger et al., 2009). Zebrafish skeletal muscle forms from the myotome compartment of the developing zebrafish somite (Holley and Nusslein-Volhard, 2000). Prior to myoblast differentiation, a nascent boundary between the somites forms and ECM is deposited at regular intervals adjacent to the notochord (Raeker and Russell, 2011). Slow skeletal muscle myoblasts elongate, extending between the anterior and posterior somite boundaries. After elongation, the myocyte begins to differentiate, assembling new myofibrils and establishing a mature cellular structure. As the slow skeletal muscle cells migrate from their position adjacent to the notochord to the periphery of the somite, they stimulate differentiation of the fast skeletal muscle that will comprise the bulk of the myotome (Henry and Amacher, 2004). The somites first form at 10 hours post fertilization (hpf) and develop in an anterior to posterior direction at a rate of one every 20–30 min until 30–34 pairs have formed (Waterman, 1969). More mature somites are therefore located in the trunk as the new somites form in the elongating tail.
Formation of the myotendinous junction
At the outset of myocyte differentiation and skeletal myofibrillogenesis in the zebrafish embryo, ECM deposition divides the developing mesoderm into distinct somites (Holley and Nusslein-Volhard, 2000). The ECM, which is enriched in fibronectin, is deposited at defined intervals along the notochord with extension of the matrix laterally and caudally to yield a “chevron-shaped” somite bounded by organized ECM (Raeker and Russell, 2011). The accumulation of ECM at the somite boundary requires a complex interplay of Eph-Ephrin signaling and α5-integrin-mediated matrix reorganization (Durbin et al., 1998). Eph-Ephrin signaling helps to confer myocyte polarity and specifies matrix deposition in defined domains. Integrins then mediate attachment of the cells to fibronectin within the somite boundary with the aid of focal adhesion kinase (FAK) (Crawford et al., 2003; Koshida et al., 2005; Julich et al., 2009). Loss of either α5-integrin or fibronectin destabilizes the somite boundary.
Elongating myoblasts attach to the matrix at the somite boundary and initiate new myofibril assembly at the site of attachment of the nascent myofibril to adhesion complexes within the sarcolemma [(Raeker and Russell, 2011) and Figure 1]. The assembly of new myofibrils is associated with progressive deposition of electron-dense material at the attachment site (Figures 1F,G) at the terminal ends of the myocyte, the zebrafish equivalent of the myotendinous junction (MTJ). As the somite boundary matures, fibronectin in the ECM is replaced by laminin and the cell-ECM contact sites become increasingly complex with the recruitment of talin, vinculin, and the dystrophin-dystroglycan complex (DGC) (Raeker and Russell, 2011).
Figure 1.
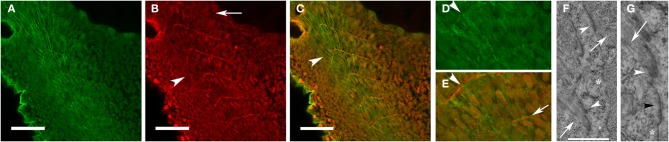
Fibronectin organization at the somite boundary precedes the assembly and organization of new myofibrils. Zebrafish embryos at 24 hpf were immunostained for fibronectin (red: B,C,E) and α-actinin (green: A,C,D). Fibronectin was organized at the somite boundary before the assembly of the first striated myofibrils (B,C: arrowheads) and becomes less abundant in the more mature boundaries (B: arrow). In the most recently formed somites, nascent myofibrils (D,E), with a characteristic irregularly dotted pattern of α-actinin staining, extend between the recently formed fibronectin-containing nascent somite/myotome boundaries (E: arrow). (F) Insertion of actin filaments (arrows) into membrane-based adhesion domains at the somite boundary is associated with the accumulation of electron-dense material (arrowheads) on the inner and outer surfaces of the sarcolemma. (G) The membrane compartment at the somite boundary undergoes progressive development in preparation for myofibril assembly. Note the absence of electron dense material in an undeveloped region of the membrane (*) with deposition of some electron-dense material in a region without well-developed actin filaments (black arrowhead) followed by more accumulation of material in a region (white arrowhead) with definite insertion of well-developed actin filaments (arrow). Scale bars are 50 μm (A–C) and 2 μm (F). (D,E) Are 3 × magnifications of corresponding regions in (A,C). (G) Is a 2.5 × magnification of the image in (F).
The development and maintenance of the MTJ has been challenging to study in higher vertebrates. Studies in the zebrafish suggest that assembly and composition of the MTJ is very similar to the connections that form at the lateral sarcolemma called costameres and that disorders such as Duchenne muscular dystrophy, in which the costameric cell-ECM attachment complexes are compromised, also have defects of the MTJ that may be clinically relevant. Characterization of the sapje zebrafish, a model of Duchenne muscular dystrophy (Bassett et al., 2003), has demonstrated that dystrophin loss is associated with destabilization of the myocyte-MTJ attachment leading to detachment and cell death. Mouse models of impaired dystrophin-DGC function have likewise demonstrated structural abnormalities of the MTJ (Law and Tidball, 1993; Grady et al., 2003) and MRI studies in patients with DMD have demonstrated preferential myocyte loss and replacement fibrosis at the termination of muscle fibers at the MTJs (Nagao et al., 1991). Therefore, modeling of MTJ assembly and stability in the zebrafish may yield important insights into disease manifestations in patients with myopathies and muscular dystrophies, particularly those that involve the dystrophin-DGC.
Relationship of the lateral sarcolemma to the underlying myofibrils
Costameres, the rib-like specialized membrane domains localized over the Z disks of the superficial myofibrils, were first described in the early 1980's (Craig and Pardo, 1983). Components of the costamere also overlying the M band but the structures are less consistent and less robust. It has been determined that the costameres are important sites for the lateral transmission of force generated by the underlying myofibrils and loss or damage of elements of the costamere, including the DGC, can lead to a range of muscular dystrophies [reviewed in Ervasti (2003)]. Yet, despite their importance, how these specialized membrane domains are developed and maintained with respect to the myofibril is not well understood. Much of what is known has been characterized in invertebrate (D. melanogaster and C. elegans) models [reviewed in Moerman and Williams (2006)]. Genetic screens for disordered sarcomere assembly and organization in C. elegans embryos led to the identification of mutants with a broad range of disorders affecting muscle structure and function. One class of mutants, the Pat (Paralyzed, Arrested elongation at Two-fold) embryonic-lethal developmental arrest mutants, were noted to have very early defects in myofibril assembly and were incapable of movement (hence the term, “paralyzed”). A second class of mutants, the Unc (Uncoordinated) mutants, is characterized by slow and/or irregular movements. Careful analysis of the mutants revealed a hierarchy of myofibrillogenesis that begins with the assembly of cell-matrix attachment complexes. Mutation or depletion of components of the transmembrane adhesion complexes, including α-integrin (Pat-2), β-integrin (Pat-3), integrin-linked kinase (Pat-4), and actopaxin (Pat-6), lead to a Pat or paralyzed phenotype in C. elegans and to comparable defects in D. melanogaster (MacKrell et al., 1988; Volk et al., 1990; Sparrow and Schock, 2009). The membrane-bound complexes organize the underlying ECM, including perlecan (Unc-52), and served as a scaffold for the assembly of the myofilaments, including the components of the sarcomere and the cellular cytoskeleton (such as kindlin/Unc-112, obscurin/Unc-89, and PINCH/Unc-97). Assembly and organization of new myofibrils progressed from the sub-sarcolemmal region to the cell interior.
However, it has been difficult to determine if a similar process occurs or is required in vertebrates. As previously noted (Kim and Ingham, 2009), new myofibril assembly in zebrafish skeletal muscle occurs in close apposition to the sarcolemma. In the zebrafish, new myofibrils are initiated at the terminal ends of the myocyte, in the sub-membrane compartment adjacent to the lateral sarcolemma of the elongated myocyte (Figure 2). Z disks of new myofibrils from adjacent myocytes were not in precise alignment but maintained a relatively consistent relationship such that the offset placed the Z disks from the myofibrils in one myocyte at approximately the A-I junction of the myofibrils in the adjacent cell [(Raeker and Russell, 2011) and Figure 2E]. The distance between the superficial myofibrils in adjacent myocytes was also maintained, suggesting that, very early in myofibril assembly, a cytoskeletal support network maintains relationships between the myofibril and specialized membrane domains that mediate attachment to the ECM (Figure 2E). The importance of the lateral membrane compartment and the submembrane cytoskeleton in scaffolding the formation of new myofibrils is supported by the preferential clustering of the cell's first myofibrils along the line of coaptation between three adjacent skeletal myocytes (Figure 2F).
Figure 2.
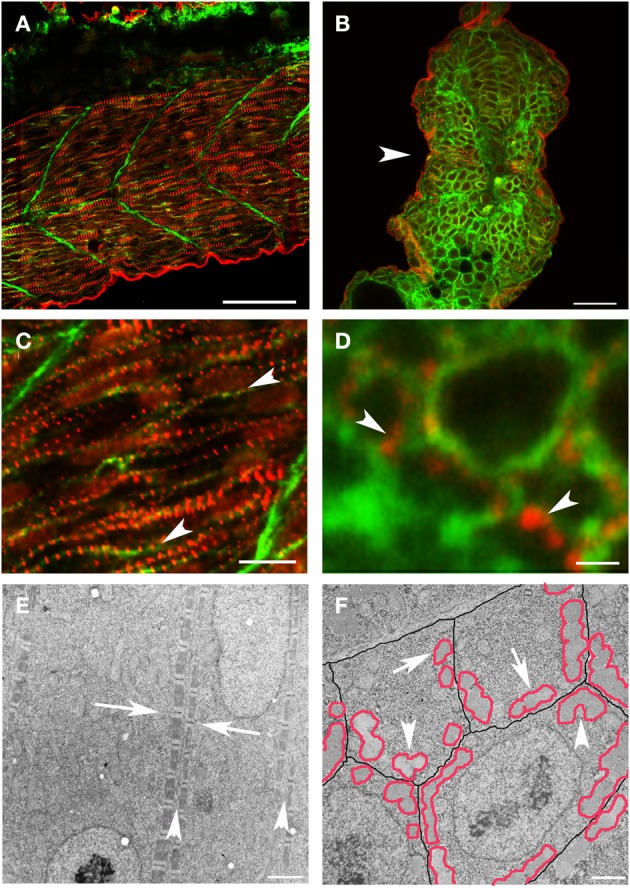
New myofibril assembly occurs adjacent to the sarcolemma in skeletal muscle. (A–D) Zebrafish embryos at 24 hpf were injected with lyn-GFP mRNA (which encodes for a GFP-fluorescent tagged protein that localizes to the cell membrane) and immunostained for α-actinin (red) and GFP (green) to mark the sarcomere and the sarcolemma respectively. Note that α-actinin dots (C,D: arrowheads) appear in a striated pattern immediately adjacent to the sarcolemma in both longitudinal optical (A,C) and transverse (B,D) sections. (E) Ultrastructural analysis reveals that there is a consistent relationship between new myofibrils that are forming on either side of the sarcolemma (E: arrowheads) and that there is a slight offset in the alignment of Z bands (E: arrows) across myocytes at this stage of development. In transverse sections (F), note that the myofibrils (outlined in red) form in close proximity to the sarcolemma (outlined in black) with preferential clustering in regions where three cells intersect (arrowheads). There are examples of myofibrils (arrows) that are not paired with a myofibril in an adjoining cell, suggesting that the organization of myofibrils across myocytes may not be direct and therefore may require patterning of the extracellular matrix to transmit architectural information. Scale bars are 50 μm (A), 20 μm (B), 10 μm (C), 1 μm (D), 2 μm (E), and 3 μm (F).
Reciprocal patterning of lateral membrane domains and newly assembled myofibrils
To further characterize the relationship between the new myofibrils and the overlying cell membrane, we examined the organization of the sarcolemma into specialized domains during early myofibril assembly. Based on previous studies (Sparrow and Schock, 2009) and the order of organization of the MTJ, we postulated that integrins would have an early and primary role in organizing the membrane domains overlying the myofibril. Therefore, we performed immunolabeling of β1-integrin during zebrafish myofibrillogenesis. We noted that early during myoblast differentiation, β1-integrin localized to linear stripes along the long axis of the myocyte (Figures 3A–C). The staining was initially continuous but rapidly transitioned to a striated pattern that closely coincided with the organization of α-actinin into nascent Z disks (Figures 3D–G).
Figure 3.
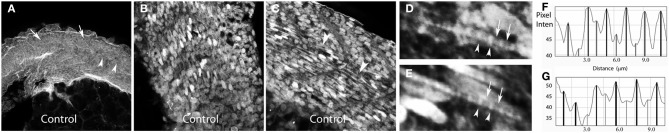
β1-integrin redistribution occurs very early in myofibrillogenesis to pattern membrane domains overlying new myofibrils. Zebrafish embryos at 20 (A) and 24 (B–G) hpf were immunolabeled for β1-integrin. Embryos demonstrated clustering of β1-integrin at new somite boundaries (arrows: A) with subsequent localization to membrane domains overlying new myofibrils in the more mature somites (arrowheads: A). (B,C) At the onset of myofibrillogenesis, β1-integrin localizes diffusely overlying the nascent myofibril before achieving a striated appearance with accumulation overlying the Z disk (arrowheads: D,E). Frequently a lower abundance and more diffuse accumulation of β1-integrin is noted at the mid A band (arrows: D,E). Line scans assessing pixel intensity notes a regular pattern of β1-integrin accumulation overlying the Z disks (black bars: F,G) with frequent smaller accumulations in the mid (A) band (grey bars: F,G). Note that, in general, the distance between Z disk accumulations of β1-integrin is less between Z disks without an intervening mid (A) band accumulation (F: far right; G: far left). Scale bars are 50 μm (A), 20 μm (B,C) and 5 μm (D,E).
As with the MTJ, these immature cell-ECM contacts may recruit other components of the adhesion complex, including talin, vinculin, and dystrophin-DGC, to form a stronger and more permanent connection. It is likely that the early connection, as evidenced by the initially diffuse localization of β1-integrin, is fluid, allowing reciprocal interactions between the membrane contact and the underlying sarcomere to coordinate the patterning of both. As development proceeded, the sarcolemma became more closely associated with the myofibril at the level of the Z disk (Raeker and Russell, 2011). Combined with the slight offset of Z disks in adjoining myocytes, this gave the sarcolemma an undulated appearance (Figure 4). This ultrastructural change was noted prior to full maturation of the costamere as elements of the DGC, while evident at the somite boundary, had not yet localized to the lateral sarcolemma (Raeker and Russell, 2011). This suggests that fixation of the myofibril to the submembrane cytoskeleton, membrane-bound cytoskeletal and cell-matrix adhesion complexes occur prior to the maturation of the DGC and may scaffold its later assembly.
Figure 4.
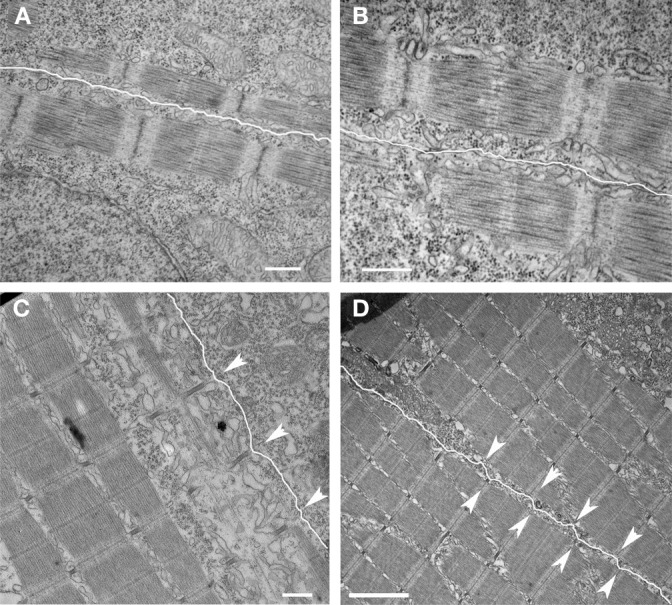
Development of nascent costameres. Skeletal muscle from zebrafish embryos at 20 hpf (A,B) demonstrate a consistent offset in the alignment of the Z disks across adjacent myocytes (sarcolemma highlighted in white). Note that the distance from the myofibril to the membrane is consistent over the length of the myofibril. By 72 hpf (C,D), the membrane has a more undulated appearance and is closest to the myofibril overlying the Z disks (arrowheads) consistent with the maturation of costameric connections from the myofibril to the sarcolemma. Scale bars are 500 nm (A–C) and 2 μm (D).
These findings suggest that specialization of the lateral sarcolemma overlying the superficial Z disks occurs very early in zebrafish myofibrillogenesis with specific localization of adhesion proteins and signaling molecules. The importance of the integrin-fibronectin attachments in anchoring the myofibril to the ECM was demonstrated in zebrafish embryos depleted of Tgfbi, an ECM protein expressed in response to TGF-beta signaling that binds to integrins (Kim and Ingham, 2009). Skeletal muscle depleted of Tgfbi formed normal appearing MTJs and myofibrils assembled in the subsarcolemmal space but the myofibrils did not remain closely attached to the sarcolemma and there was markedly reduced myofibril content. Similarly, zebrafish depleted of fibronectin demonstrated reduced myofibril content and organization (Snow et al., 2008). These studies suggest that integrin-mediated cell-ECM adhesion is an important stimulus for hypertrophic growth and an important regulator of myofibril organization but is not essential for myofibril assembly in zebrafish skeletal muscle in vivo.
The involvement of the ECM in the organization of myofibrils within the cell (Russell et al., 2000; Grosberg et al., 2011) may account for the coordination of the assembly process across adjacent cells resulting in the alignment of Z disks in register from one myocyte to the next. Alignment of sarcomeres across myocytes has been noted previously both in heart and skeletal muscle and is thought to have a role in optimizing force transfer and contractile response. Pathologic disease states such as end-stage heart failure (Schaper et al., 1991; Sharov et al., 1994), hypertrophic cardiomyopathy (Ferrans et al., 1972) and many myopathies (Milner et al., 1996; Marbini et al., 1998) and muscular dystrophies are all characterized by myofibril disarray and abnormal relationships between myocytes. The alignment process has not been well defined but has been attributed to patterning of the ECM by the myocyte (Caulfield and Borg, 1979; Robinson et al., 1987) or to direct patterning of adjacent cells by cell-cell contacts.
Remodeling of myofibrillar clusters as adjacent myofibrils are bundled into larger units
Within a given myocyte, as the peripheral myofibrillar clusters joined to form larger functional units, there was extensive remodeling of the sarcomere structure. If the adjacent myofibrils were slightly offset, the sarcomere structure remained intact, suggesting some capacity for myofibrils to “slide” into alignment during the bundling process. However, there were occasionally noted to be regions in which the offset between the Z disks of fusing myofibrils appeared to be too great to permit intact integration. In those regions, there were noted to be zones of transition in which existing myofibrils were dismantled and then reassembled. This was evident from the absence of intact sarcomeres in the transition zone between the part of the myofibril that had been incorporated into the adjoining myofibril and the part that was still unbundled (Figure 5). This was also noted on cross-sections where, in the areas of myofibrillar fusion, there was loss of the hexagonal arrangement of the actin and myosin filaments suggesting remodeling of the sarcomere's cytoskeletal support network in the transition zone.
Figure 5.

Lateral integration of new myofibrils involves coordinate remodeling of the sarcomeres, the cellular cytoskeleton and the surrounding membranes. (A) New skeletal myofibrils in a 20 hpf zebrafish embryo undergoing lateral fusion show progressive alignment at the level of the Z disks (arrowheads). Note the paucity of myofilaments in the intervening region between the Z disk involved in the active lateral incorporation (arrow) with intact sarcomeres on either end of the remodeling sarcomere. (B) A higher magnification of the image in (A) demonstrating the area of remodeling with interruption of the myosin filaments (arrowhead). (C) Newly assembled sarcomeres demonstrating recent alignment of the Z disks in register one end of the sarcomere (arrow) and misalignment at the other end (arrowhead). (D) Lateral integration of myofibrils with loss of defined sarcomere structure of the fusing myofibril (arrow) with progressive organization of the Z disk (arrowhead). (E) Higher magnification image of the image in (D) demonstrating remodeling of the T tubule/sarcoplasmic reticulum with loss of the membranes (white arrow) as the myofibrils fuse (black arrow). Note the disturbance of sarcomere architecture as the myofibrils fuse (arrowhead). (F) Transverse sections reveal that, as myofibrils (M1 and M2) fuse, the sarcoplasmic reticulum (arrowhead) withdraws from the line of apposition, the hexagonal array of myofilaments is disordered (*) on either side of the interface suggesting remodeling of the supporting cytoskeleton. Note that the new myofibrils are “separated” from the rest of the sarcoplasm by the membranes of the SR. Scale bars are 500 nm (A–E) and 100 nm (F).
One interesting feature of myofibril assembly and organization is how dynamic the process appears to be. Previous studies by the Sanger laboratory have revealed that sarcomeres are very dynamic structures in which there is a continuous protein turnover, even in intact, mature myofibrils (Sanger et al., 2005; Wang et al., 2005; Stout et al., 2008). Prior characterization of the myofibril organization and alignment process in primary cultures of chick skeletal myotubes in which Z disk alignment was described to progress by a “trial and error” that involved the disassembly of existing myofibrillar structures (McKenna et al., 1986). Proteosomal complexes strongly co-localize with new myofibrils (Foucrier et al., 1999, 2001) and mice lacking calpain-3, a cysteine protease, had significant abnormalities of sarcomere organization (Kramerova et al., 2004). Previous work from the invertebrate C. elegans model supports a role for ubiquitin-mediated proteolysis in sarcomere protein turnover with demonstration that (i) CSN-5, a component of the COP9 signalsome regulates levels of selected sarcomeric proteins (Miller et al., 2009) and (ii) MEL-26, an adapter that links cullin-3/ubuiquitin ligase complexes to the giant sarcomeric protein Unc-89, regulates thick filament organization (Wilson et al., 2012). The balance between chaperone-mediated protein assembly and ubiquitin-mediated protein degradation appears to have a central role in the assembly and remodeling of the myofibril in invertebrate models [reviewed in Willis et al. (2009); Kim et al. (2008)]. Our findings in the zebrafish support important role of protein complex assembly and degradation in myofibril organization and re-organization during development and suggest that the zebrafish may be an excellent model in which to study these dynamic processes.
Organization of the membrane compartments surrounding the myofibril
Myocyte organization extends beyond the myofibril and sarcolemma to involve the internal membrane compartments involved in excitation-contraction coupling. Muscle contraction depends on the release and re-uptake of calcium stores from specialized membrane compartments to initiate contraction and permit relaxation respectively. Like the other elements of the myocyte, these membrane compartments, which include the junctional sarcoplasmic reticulum and invaginations of the sarcolemma called T tubules, are organized with respect to the underlying myofibril. As with other elements of the myocyte organization process, how these membrane compartments link to and become precisely situated over specific sarcomere domains is not well understood. Since these structures only exist in vertebrate muscle and the timing of their appearance during development varies between vertebrate species and between cardiac and skeletal myocytes, it has been very difficult to identify the structural elements required for their formation.
We used electron microscopic analysis to characterize the formation of the T tubules and sarcoplasmic reticulum during development. Unlike mammalian cardiac myocytes in which mature T tubules are frequently not noted until after birth, T tubules were noted, albeit not consistently, at the level of the Z disk in the newly formed myofibrils shortly after the appearance of mature sarcomeres (Figure 6F). T tubules appeared first as finger-like invaginations of the sarcolemma and later increased in cross-sectional area (Figures 6A–E). The sarcoplasmic reticulum was first diffusely localized in subsarcolemmal space and appeared more specialized on the cytoplasmic side of new myofibrils where junctional domains were noted associated with the T tubules. As was noted with the myofibrils, extensive remodeling of the T tubules and sarcoplasmic reticulum were noted in regions where adjacent myofibrils were bundling into larger units. Taken together with the findings from previous studies, our findings suggest that T tubular architecture is directly dependent on the cytoskeletal support network for the myofibril, and that the components of the cytoskeleton that are directly involved in anchoring the transverse domains of the T tubules are closely related to and may participate in the transverse alignment of myofibrils in register. Furthermore, the early formation of the T tubules in zebrafish skeletal muscle suggests that, even in species and tissues where T tubules form much later in development, the cytoskeletal network required to support T tubules may be present very early in new myofibril assembly. This network reorganizes to permit the lateral integration of myofibrils during new myofibril assembly and during adaptive cellular remodeling, leading to a loss of the transverse arrangement of the T tubules during these processes. Linkage of the T tubule system to the cytoskeleton may occur through spectrin (Messina and Lemanski, 1991), a sub-membrane cytoskeletal protein, that directly interacts with membrane-bound elements including ankyrin (Bennett and Stenbuck, 1979). Interaction between ankyrin and spectrin may help form a site of membrane anchorage to help nucleate the assembly of new myofibrils (Bennett et al., 2004) and contribute to the lattice that supports the T tubules (Messina and Lemanski, 1991). It has been postulated that ankyrin within the sarcolemma and the subsarcolemmal spectrin cytoskeleton are involved in establishing the membrane microdomains overlying the superficial myofibrils (Bennett et al., 2004).
Figure 6.
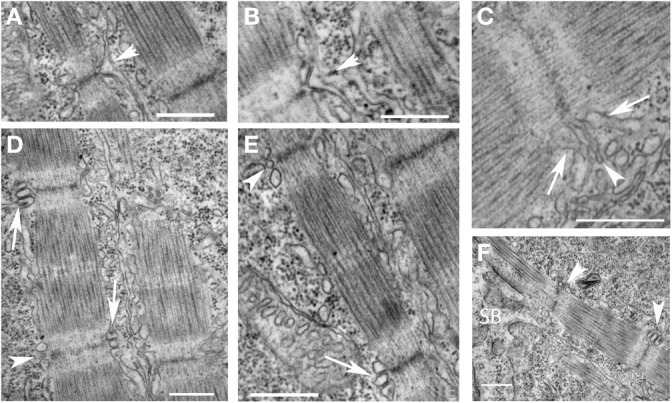
Early organization of T tubules and junctional SR around newly-formed myofibrils. (A,B) T tubules formed from invaginations (arrowheads) of the sarcolemma overlying the Z disks of newly formed myofibrils. (C) The T tubules (arrowhead) were often initially quite narrow initially before increasing in volume later in development. The sarcoplasmic reticulum (arrows) organized on either side of invaginating T tubules but did not initially appear to be in direct contact. (D) Triads (two junctional SR domains surrounding a T tubule) formed overlying the Z disk of newly formed myofibrils progressing from immature structures with small, round T tubules (arrowheads) to more mature structures with larger, oval T tubules in close apposition to the junctional SR (arrows). (E) A higher magnification image of a new myofibril with an immature (arrowhead) and more mature (arrow) triad at either end of the sarcomere. (F) Triads (arrowheads) develop around newly formed Z disks as new sarcomeres are added adjacent to the somite boundary (SB). Scale bars are 500 nm.
Modeling of human myopathies and muscular dystrophies
The ability to study dynamic relationships between adjacent myofibrils and between myofibrils and the surrounding membrane domains has encouraged the increased utilization of the zebrafish model for the understanding of the pathogenesis human myopathies and muscular dystrophies. Several muscular dystrophy mutant strains of zebrafish have been identified including those with mutations in dystrophin (sapje), laminin α2 (candyfloss), and dystroglycan (pachytail and dag1hu3072) (Bassett et al., 2003; Hall et al., 2007; Gupta et al., 2011) while several other genes causing muscular dystrophy have been disrupted by antisense morpholino approaches [reviewed in Gibbs et al. (2013a)]. One key phenotypic feature of many muscular dystrophy mutant zebrafish strains is disturbed birefringence of the skeletal muscle. The transparency of the zebrafish embryo enables direct visualization of skeletal muscle under polarized light. In wild-type embryos, the near crystalline structure of the ordered sarcomeres results in the unequal refraction of polarized light as it passes through skeletal muscle of the zebrafish trunk and tail, a property called birefringence. The intensity of the birefringence is reduced in the settings of myopathies and muscular dystrophies in which the myofibrillar architecture is disturbed. Therefore, measurements of the birefringence of the skeletal muscle of zebrafish can be used to assess the severity of myopathic changes in models of myopathy and muscular dystrophy, enabling the high-throughput assessment of disease severity and response to treatment.
In addition to myofibrillar structural integrity, muscular dystrophy mutant zebrafish also show marked changes in sarcolemmal integrity, an important hallmark of many muscular dystrophies. Loss of sarcolemma integrity has been demonstrated in DGC-deficient and in laminin beta2-deficient zebrafish models (Hall et al., 2007; Jacoby et al., 2009; Lin et al., 2011). The importance of sarcolemma integrity in muscle fiber maintenance is supported by the findings that morpholino disruption of one of the key proteins in the muscle membrane repair pathway, dysferlin, loss of which causes LGMD 2B and Myoshi myopathy in humans, also causes disrupted birefringence similar to DGC-deficient dystrophic zebrafish (Kawahara et al., 2011a). Zebrafish models should provide new insights into the relatively unknown plasma membrane repair pathways in muscle, because the optical transparency of zebrafish embryos renders the whole muscle amenable to high resolution real time microscope imaging to monitor the repair process (Roostalu and Strahle, 2012).
Zebrafish are becoming an increasingly important tool for screening for new therapeutic compounds. The validity and feasibility of performing high throughput screens for muscular dystrophy using zebrafish is supported by recent work of Kunkel and colleagues, who used birefringence to screen the Prestwick chemical library for compounds that restored muscle architecture in a zebrafish DMD model (Kawahara et al., 2011b). Future screens in zebrafish that utilize functional outputs such as swimming behaviors, as described by Kuwada and colleagues (Hirata et al., 2004; Horstick et al., 2013), may yield important new targets for therapy of muscular dystrophies that not only improve muscle structural integrity, but also improve muscle weakness and dysfunction seen in human patients that is independent of mechanical injury and loss of sarcolemma integrity (Gumerson et al., 2010, 2013).
Application of genome editing techniques for manipulation of the zebrafish genome
Targeted germline modifications in the murine genome has enabled production of both “knock-out” and “knock-in” mice, allowing the development of animal models to study a wide range of human disease processes. However, gene targeting in the mouse can be expensive and time-consuming and the murine model may not be as amenable as the zebrafish to study dynamic processes that occur during development. Yet, until recently, the targeting of specific genes in zebrafish for the creation of disease models (i) was limited to morpholino antisense RNA-mediated inhibition of gene expression (which can cause non-specific toxic effects on development) or (ii) were the result of large, random mutagenesis screens where the strain harboring the mutation in the gene of interest needed to be identified by an exhaustive screening or specific phenotype manifestation. More recently, genome editing nucleases have been identified and used to help create specific disease models in the zebrafish. These nucleases have the capability to bind a specific DNA sequence and induce a double-stranded DNA break at that site. The inclusion of a homologous DNA fragment facilitates recombination and thus “edits” the genomic locus. In the absence of a repair template, non-homologous end-joining occurs, resulting in insertions or deletions, and subsequent frameshift mutations and/or truncations. In the presence of a repair template, the targeted DNA sequence can be replaced by one that contains a specific mutation, allowing the introduction of known or suspected disease-causing mutations into specific functional domains. There are two gene editing strategies, TALENs and CRISPR, that have emerged. TALENs (transcription activator-like effector nucleases) use DNA binding domains derived from a plant pathogen (Joung and Sander, 2013) and have been shown to be efficient at producing knockouts (Huang et al., 2011; Sander et al., 2011) and in vivo homologous recombination (Bedell et al., 2012; Zu et al., 2013) in zebrafish. CRISPR (clustered regularly interspaced short palindromic repeats), which works by a slightly different mechanism, is a set of genomic repeats found in bacteria and archaea that function as a system of adaptive immunity (Wiedenheft et al., 2012). As opposed to TALENs, CRISPR recognizes target sites using short guide RNAs that anneal to complementary genomic DNA sequences and recruit the Cas9 nuclease to effect cleavage (Blackburn et al., 2013). CRISPR has been found to be effective at generating knockouts (Hwang et al., 2013a), homologous recombination (Chang et al., 2013; Hwang et al., 2013b), and chromosomal deletions and inversions (Xiao et al., 2013). With these novel tools, rapid, efficient and inexpensive manipulation of the zebrafish genome can now be performed to create models of human disease.
Use of the zebrafish for the identification of novel therapies
The high degree of molecular and structural identity, combined with the rapid development and large clutch size, makes zebrafish an ideal system for in vivo drug discovery to identify novel therapies. Drug discovery efforts can be largely divided into two approaches: targeted compound screening evolved from hypothesis-driven experimentation and large scale drug screening. Several previous studies have highlighted the utility of the zebrafish for targeted compound screening in models of human muscle diseases. Henry and colleagues demonstrated the ability of NAD+ to prevent dystrophic changes in DGC—or integrin α7—deficient zebrafish (Goody et al., 2012). Based on studies showing abnormal neuromuscular junctions in zebrafish with model of dynamin-2 centronuclear myopathy, acetylcholinesterase inhibitors were identified as modulators of the motor phenotype (Gibbs et al., 2013b). These examples reveal how the zebrafish model can serve as a quick and potent tool for testing specific chemical compounds. However, the real power of the fish lies in the ability to test drug libraries in order to discovery previously unidentified molecular and pharmacologic pathways.
Large scale drug screens have previously been not feasible in vivo in vertebrate models of human muscle disease. Now, however, with the characterization of numerous mutants in the zebrafish that mirror the genetics and the cellular abnormalities of their human disease counterparts, such screens have become a reality. This is particularly aided by the facts that zebrafish develop rapidly and ex utero, can be produced in large numbers, and readily absorb drugs presented in the media. Several screens have been successfully performed in models of a range of human diseases. The most high profile to date has been a screen by Zon and colleagues for melanoma, as they identified compounds using a zebrafish screen that are now in clinical trials for patients with melanoma (White et al., 2011). A single large scale screen has been completed and published for a zebrafish model of Duchenne Muscular Dystrophy (the sapje zebrafish) by Kunkel and colleagues (Kawahara et al., 2011b). The screen utilized birefringence, which is abnormal in all known zebrafish models of muscular dystrophy, as a read out. They identified 48 positive “hits” that prevented the abnormal birefringence in the sapje model, a list that included both known and newly identified drug targets for DMD. Subsequent analyses by this group based on their screen have focused on refining drug discovery related to cAMP-dependent protein kinase A modulation. Their results have laid the groundwork for future screens by (i) identifying known targets and thus validating the fish as a relevant model for screening drugs and (ii) discovering new target pathways and thus demonstrating the utility and power of zebrafish-based drug discovery studies.
Summary
The developing zebrafish embryo has many properties that make it a very powerful model to study myofibril assembly and myocyte organization. Work in our laboratory and others suggest that there is a concurrent maturation and organization of the myofibrils and membrane compartments involved in lateral and longitudinal force transmission (the costameres and MTJs, respectively) and those involved in excitation-contraction coupling (the T tubules and sarcoplasmic reticulum) (Figure 7). The process appears to be dynamic and iterative, as assembled structures are disassembled and reorganized to align with other myofibrillar structures within the cell and within neighboring cells. Interactions between the sarcomere and the ECM help to determine tissue organization ensuring parallel transmission of contractile forces and resulting in optimal muscle performance. This work supports the characterization of sarcomeric, sarcolemmal, cytoskeletal and ECM remodeling as fully integrated processes during muscle development. As such, it will be important to characterize the response of each in the adaptive response to muscle injury or myopathy. Therapeutic targeting of the elements responsible for coordinating the relationships between these cellular compartments may promote preservation of muscle function or promote muscle repair in response to injury. The application of gene editing approaches and high throughput drug discovery and therapeutic testing will make the zebrafish an important resource in understanding the pathophysiology of a broad range of muscle disorders and in the identification of novel therapeutic strategies.
Figure 7.
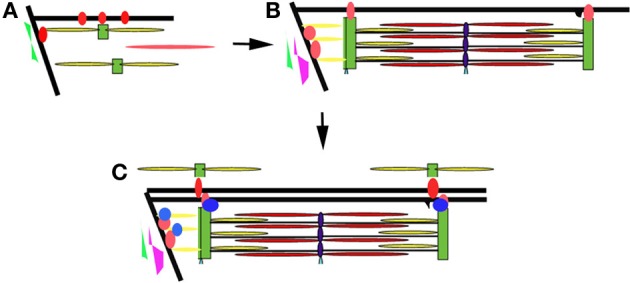
Model of myofibril assembly and myocyte organization. (A) The first myofibrils form at the insertion of actin filaments (yellow rods) and myosin filaments (red rod) at the terminal ends of the myocyte along the lateral sarcolemma. Integrins (red circles) are initially diffusely organized along the sarcolemma before concentrating at sites of actin insertion sites (terminal ends of the myofibril and overlying the Z disks of the superficial myofibrils) where they help organize the surrounding ECM including fibronectin (in green). (B) As myofibrillogenesis proceeds, integrins become more concentrated at contact sites and laminin (purple) is deposited along the somite border. Sarcolemmal invaginations that will eventually become the T tubular network form overlying the Z disks (black projection). (C) Additional adhesion complex proteins (blue) are recruited to stabilize and strengthen the contacts at the terminal ends of the myofibril. The patterning of the sarcolemma and the surrounding ECM results in the patterning of the sarcolemma and subsequently the newly assembling myofibrils of neighboring cells.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Ashley Bieniek for her assistance with the care of the zebrafish. The work was supported by funds from the Aaron Stern Professorship (Mark W. Russell) and the NIH (HL075093). We would like to thank Dr. Scott Holley and Dr. Robert Bloch for helpful comments.
Glossary
Abbreviations
- SR
Sarcoplasmic reticulum
- GFP
green fluorescent protein
- ECM
extracellular matrix
- TGF
transforming growth factor.
References
- Bassett D. I., Bryson-Richardson R. J., Daggett D. F., Gautier P., Keenan D. G., Currie P. D. (2003). Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development 130, 5851–5860 10.1242/dev.00799 [DOI] [PubMed] [Google Scholar]
- Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., Krug R. G., et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 10.1038/nature11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Baines A. J., Lecomte M. C., Maggs A. M., Pinder J. C. (2004). Not just a plasma membrane protein: in cardiac muscle cells alpha-II spectrin also shows a close association with myofibrils. Muscle Res. Cell Motil. J. 25, 119–126 10.1023/B:JURE.0000035892.77399.51 [DOI] [PubMed] [Google Scholar]
- Bennett V., Stenbuck P. J. (1979). Identification and partial purification of ankyrin, the high affinity membrane attachment site for human erythrocyte spectrin. Biol. Chem. J. 254, 2533–2541 [PubMed] [Google Scholar]
- Blackburn P. R., Campbell J. M., Clark K. J., Ekker S. C. (2013). The CRISPR system–keeping zebrafish gene targeting fresh. Zebrafish 10, 116–118 10.1089/zeb.2013.9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capetanaki Y., Bloch R. J., Kouloumenta A., Mavroidis M., Psarras S. (2007). Muscle intermediate filaments and their links to membranes and membranous organelles. Exp. Cell Res. 313, 2063–2076 10.1016/j.yexcr.2007.03.033 [DOI] [PubMed] [Google Scholar]
- Caulfield J. B., Borg T. K. (1979). The collagen network of the heart. Lab. Invest. 40, 364–372 [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig S. W., Pardo J. V. (1983). Gamma actin, spectrin, and intermediate filament proteins colocalize with vinculin at costameres, myofibril-to-sarcolemma attachment sites. Cell Motil. 3, 449–462 10.1002/cm.970030513 [DOI] [PubMed] [Google Scholar]
- Crawford B. D., Henry C. A., Clason T. A., Becker A. L., Hille M. B. (2003). Activity and distribution of paxillin, focal adhesion kinase, and cadherin indicate cooperative roles during zebrafish morphogenesis. Mol. Biol. Cell 14, 3065–3081 10.1091/mbc.E02-08-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin L., Brennan C., Shiomi K., Cooke J., Barrios A., Shanmugalingam S., et al. (1998). Eph signaling is required for segmentation and differentiation of the somites. Genes Dev. 12, 3096–3109 10.1101/gad.12.19.3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti J. M. (2003). Costameres: the Achilles' heel of Herculean muscle. Biol. Chem. J. 278, 13591–13594 10.1074/jbc.R200021200 [DOI] [PubMed] [Google Scholar]
- Ferrans V. J., Morrow A. G., Roberts W. C. (1972). Myocardial ultrastructure in idiopathic hypertrophic subaortic stenosis. A study of operatively excised left ventricular outflow tract muscle in 14 patients. Circulation 45, 769–792 10.1161/01.CIR.45.4.769 [DOI] [PubMed] [Google Scholar]
- Foucrier J., Bassaglia Y., Grand M. C., Rothen B., Perriard J. C., Scherrer K. (2001). Prosomes form sarcomere-like banding patterns in skeletal, cardiac, and smooth muscle cells. Exp. Cell Res. 266, 193–200 10.1006/excr.2001.5212 [DOI] [PubMed] [Google Scholar]
- Foucrier J., Grand M. C., De Conto F., Bassaglia Y., Geraud G., Scherrer K., et al. (1999). Dynamic distribution and formation of a para-sarcomeric banding pattern of prosomes during myogenic differentiation of satellite cells in vitro. Cell Sci. J. 112 (Pt 7), 989–1001 [DOI] [PubMed] [Google Scholar]
- Gibbs E. M., Horstick E. J., Dowling J. J. (2013a). Swimming into prominence: the zebrafish as a valuable tool for studying human myopathies and muscular dystrophies. FEBS J. 280, 4187–4197 10.1111/febs.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs E. M., Clarke N. F., Rose K., Oates E. C., Webster R., Feldman E. L., et al. (2013b). Neuromuscular junction abnormalities in DNM2-related centronuclear myopathy. Mol. Med. J. 91, 727–737 10.1007/s00109-013-0994-4 [DOI] [PubMed] [Google Scholar]
- Goody M. F., Kelly M. W., Reynolds C. J., Khalil A., Crawford B. D., Henry C. A. (2012). NAD+ biosynthesis ameliorates a zebrafish model of muscular dystrophy. PLoS Biol. 10:e1001409 10.1371/journal.pbio.1001409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady R. M., Akaaboune M., Cohen A. L., Maimone M. M., Lichtman J. W., Sanes J. R. (2003). Tyrosine-phosphorylated and nonphosphorylated isoforms of alpha-dystrobrevin: roles in skeletal muscle and its neuromuscular and myotendinous junctions. Cell Biol. J. 160, 741–752 10.1083/jcb.200209045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg A., Kuo P. L., Guo C. L., Geisse N. A., Bray M. A., Adams W. J., et al. (2011). Self-organization of muscle cell structure and function. PLoS Comput. Biol. 7:e1001088 10.1371/journal.pcbi.1001088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumerson J. D., Davis C. S., Kabaeva Z. T., Hayes J. M., Brooks S. V., Michele D. E. (2013). Muscle-specific expression of LARGE restores neuromuscular transmission deficits in dystrophic LARGE(myd) mice. Hum. Mol. Genet. 22, 757–768 10.1093/hmg/dds483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumerson J. D., Kabaeva Z. T., Davis C. S., Faulkner J. A., Michele D. E. (2010). Soleus muscle in glycosylation-deficient muscular dystrophy is protected from contraction-induced injury. Am. Physiol. Cell Physiol. J. 299, C1430–C1440 10.1152/ajpcell.00192.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Kawahara G., Gundry S. R., Chen A. T., Lencer W. I., Zhou Y., et al. (2011). The zebrafish dag1 mutant: a novel genetic model for dystroglycanopathies. Hum. Mol. Genet. 20, 1712–1725 10.1093/hmg/ddr047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. E., Bryson-Richardson R. J., Berger S., Jacoby A. S., Cole N. J., Hollway G. E., et al. (2007). The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. U.S.A. 104, 7092–7097 10.1073/pnas.0700942104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein S., Kostin S., Heling A., Maeno Y., Schaper J. (2000). The role of the cytoskeleton in heart failure. Cardiovasc. Res. 45, 273–278 10.1016/S0008-6363(99)00268-0 [DOI] [PubMed] [Google Scholar]
- Henry C. A., Amacher S. L. (2004). Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Dev. Cell 7, 917–923 10.1016/j.devcel.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Hirata H., Saint-Amant L., Waterbury J., Cui W., Zhou W., Li Q., et al. (2004). accordion, a zebrafish behavioral mutant, has a muscle relaxation defect due to a mutation in the ATPase Ca2+ pump SERCA1. Development 131, 5457–5468 10.1242/dev.01410 [DOI] [PubMed] [Google Scholar]
- Holley S. A., Nusslein-Volhard C. (2000). Somitogenesis in zebrafish. Curr. Top. Dev. Biol. 47, 247–277 [DOI] [PubMed] [Google Scholar]
- Horstick E. J., Linsley J. W., Dowling J. J., Hauser M. A., McDonald K. K., Ashley-Koch A., et al. (2013). Stac3 is a component of the excitation-contraction coupling machinery and mutated in Native American myopathy. Nat. Commun. 4, 1952 10.1038/ncomms2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Xiao A., Zhou M., Zhu Z., Lin S., Zhang B. (2011). Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Kaini P., Sander J. D., et al. (2013a). Heritable and precise zebrafish genome editing using a CRISPR-Cas System. PLoS ONE 8:e68708 10.1371/journal.pone.0068708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., et al. (2013b). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby A. S., Busch-Nentwich E., Bryson-Richardson R. J., Hall T. E., Berger J., Berger S., et al. (2009). The zebrafish dystrophic mutant softy maintains muscle fibre viability despite basement membrane rupture and muscle detachment. Development 136, 3367–3376 10.1242/dev.034561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J. K., Sander J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 10.1038/nrm3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julich D., Mould A. P., Koper E., Holley S. A. (2009). Control of extracellular matrix assembly along tissue boundaries via Integrin and Eph/Ephrin signaling. Development 136, 2913–2921 10.1242/dev.038935 [DOI] [PubMed] [Google Scholar]
- Kanagawa M., Toda T. (2006). The genetic and molecular basis of muscular dystrophy: roles of cell-matrix linkage in the pathogenesis. Hum. Genet. J. 51, 915–926 10.1007/s10038-006-0056-7 [DOI] [PubMed] [Google Scholar]
- Kawahara G., Serafini P. R., Myers J. A., Alexander M. S., Kunkel L. M. (2011a). Characterization of zebrafish dysferlin by morpholino knockdown. Biochem. Biophys. Res. Commun. 413, 358–363 10.1016/j.bbrc.2011.08.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara G., Karpf J. A., Myers J. A., Alexander M. S., Guyon J. R., Kunkel L. M. (2011b). Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. U.S.A. 108, 5331–5336 10.1073/pnas.1102116108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. D., Kim D. J., Lee I. J., Rah B. J., Sawa Y., Schaper J. (1992). Human fetal heart development after mid-term: morphometry and ultrastructural study. Mol. Cell Cardiol. J. 24, 949–965 10.1016/0022-2828(92)91862-Y [DOI] [PubMed] [Google Scholar]
- Kim H. R., Ingham P. W. (2009). The extracellular matrix protein TGFBI promotes myofibril bundling and muscle fibre growth in the zebrafish embryo. Dev. Dyn. 238, 56–65 10.1002/dvdy.21812 [DOI] [PubMed] [Google Scholar]
- Kim J., Lowe T., Hoppe T. (2008). Protein quality control gets muscle into shape. Trends Cell Biol. 18, 264–272 10.1016/j.tcb.2008.03.007 [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A., Ackermann M. A., Bowman A. L., Yap S. V., Bloch R. J. (2009). Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol. Rev. 89, 1217–1267 10.1152/physrev.00017.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshida S., Kishimoto Y., Ustumi H., Shimizu T., Furutani-Seiki M., Kondoh H., et al. (2005). Integrinalpha5-dependent fibronectin accumulation for maintenance of somite boundaries in zebrafish embryos. Dev. Cell 8, 587–598 10.1016/j.devcel.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Kramerova I., Kudryashova E., Tidball J. G., Spencer M. J. (2004). Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 13, 1373–1388 10.1093/hmg/ddh153 [DOI] [PubMed] [Google Scholar]
- Law D. J., Tidball J. G. (1993). Dystrophin deficiency is associated with myotendinous junction defects in prenecrotic and fully regenerated skeletal muscle. Am. Pathol. J. 142, 1513–1523 [PMC free article] [PubMed] [Google Scholar]
- Lin Y. Y., White R. J., Torelli S., Cirak S., Muntoni F., Stemple D. L. (2011). Zebrafish Fukutin family proteins link the unfolded protein response with dystroglycanopathies. Hum. Mol. Genet. 20, 1763–1775 10.1093/hmg/ddr059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKrell A. J., Blumberg B., Haynes S. R., Fessler J. H. (1988). The lethal myospheroid gene of Drosophila encodes a membrane protein homologous to vertebrate integrin beta subunits. Proc. Natl. Acad. Sci. U.S.A. 85, 2633–2637 10.1073/pnas.85.8.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbini A., Gemignani F., Badiali L., Bellanova M. F., Margarito F. (1998). Congenital myopathy with mosaic fibers and interlacing sarcomeres: a new structural myopathy. Acta Neuropathol. 96, 643–650 10.1007/s004010050946 [DOI] [PubMed] [Google Scholar]
- McKenna N. M., Johnson C. S., Wang Y. L. (1986). Formation and alignment of Z lines in living chick myotubes microinjected with rhodamine-labeled alpha-actinin. Cell Biol. J. 103, 2163–2171 10.1083/jcb.103.6.2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina D. A., Lemanski L. F. (1991). Studies of hamster cardiac myofibrillogenesis in vivo with antibodies to spectrin, desmin, and alpha-actinin. Am. Anat. J. 191, 85–94 10.1002/aja.1001910109 [DOI] [PubMed] [Google Scholar]
- Miller R. K., Qadota H., Stark T. J., Mercer K. B., Wortham T. S., Anyanful A., et al. (2009). CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle. Mol. Biol. Cell 20, 3608–3616 10.1091/mbc.E09-03-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner D. J., Weitzer G., Tran D., Bradley A., Capetanaki Y. (1996). Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. Cell Biol. J. 134, 1255–1270 10.1083/jcb.134.5.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D. G., Williams B. D. (2006). Sarcomere assembly in C. elegans muscle, ed The C. elegans Research Community , WormBook (WormBook) 10.1895/wormbook.1.81.1 Available online at: http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Nagao H., Morimoto T., Sano N., Takahashi M., Nagai H., Tawa R., et al. (1991). Magnetic resonance imaging of skeletal muscle in patients with Duchenne muscular dystrophy–serial axial and sagittal section studies. No To Hattatsu 23, 39–43 [PubMed] [Google Scholar]
- Raeker M. O., Russell M. W. (2011). Obscurin depletion impairs organization of skeletal muscle in developing zebrafish embryos. Biomed. Biotechnol. J. 2011, 479135 10.1155/2011/479135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee D., Sanger J. M., Sanger J. W. (1994). The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil. Cytoskeleton 28, 1–24 10.1002/cm.970280102 [DOI] [PubMed] [Google Scholar]
- Robinson T. F., Factor S. M., Capasso J. M., Wittenberg B. A., Blumenfeld O. O., Seifter S. (1987). Morphology, composition, and function of struts between cardiac myocytes of rat and hamster. Cell Tissue Res. 249, 247–255 10.1007/BF00215507 [DOI] [PubMed] [Google Scholar]
- Roostalu U., Strahle U. (2012). In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell 22, 515–529 10.1016/j.devcel.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Russell B., Motlagh D., Ashley W. W. (2000). Form follows function: how muscle shape is regulated by work. Appl. Physiol. J. 88, 1127–1132 [DOI] [PubMed] [Google Scholar]
- Sander J. D., Cade L., Khayter C., Reyon D., Peterson R. T., Joung J. K., et al. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J. W., Kang S., Siebrands C. C., Freeman N., Du A., Wang J., et al. (2005). How to build a myofibril. Muscle Res. Cell Motil. J. 26, 343–354 10.1007/s10974-005-9016-7 [DOI] [PubMed] [Google Scholar]
- Sanger J. W., Wang J., Fan Y., White J., Sanger J. M. (2010). Assembly and dynamics of myofibrils. Biomed. Biotechnol. J. 2010, 858606 10.1155/2010/858606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J. W., Wang J., Holloway B., Du A., Sanger J. M. (2009). Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell Motil. Cytoskeleton 66, 556–566 10.1002/cm.20365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper J., Froede R., Hein S., Buck A., Hashizume H., Speiser B., et al. (1991). Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation 83, 504–514 10.1161/01.CIR.83.2.504 [DOI] [PubMed] [Google Scholar]
- Sharov V. G., Sabbah H. N., Shimoyama H., Ali A. S., Levine T. B., Lesch M., et al. (1994). Abnormalities of contractile structures in viable myocytes of the failing heart. Int. Cardiol. J. 43, 287–297 10.1016/0167-5273(94)90209-7 [DOI] [PubMed] [Google Scholar]
- Snow C. J., Peterson M. T., Khalil A., Henry C. A. (2008). Muscle development is disrupted in zebrafish embryos deficient for fibronectin. Dev. Dyn. 237, 2542–2553 10.1002/dvdy.21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow J. C., Schock F. (2009). The initial steps of myofibril assembly: integrins pave the way. Nat. Rev. Mol. Cell Biol. 10, 293–298 10.1038/nrm2634 [DOI] [PubMed] [Google Scholar]
- Stout A. L., Wang J., Sanger J. M., Sanger J. W. (2008). Tracking changes in Z-band organization during myofibrillogenesis with FRET imaging. Cell Motil. Cytoskeleton 65, 353–367 10.1002/cm.20265 [DOI] [PubMed] [Google Scholar]
- Terai M., Komiyama M., Shimada Y. (1989). Myofibril assembly is linked with vinculin, alpha-actinin, and cell-substrate contacts in embryonic cardiac myocytes in vitro. Cell Motil. Cytoskeleton 12, 185–194 10.1002/cm.970120402 [DOI] [PubMed] [Google Scholar]
- Volk T., Fessler L. I., Fessler J. H. (1990). A role for integrin in the formation of sarcomeric cytoarchitecture. Cell 63, 525–536 10.1016/0092-8674(90)90449-O [DOI] [PubMed] [Google Scholar]
- Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J. M., et al. (2005). Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskeleton 61, 34–48 10.1002/cm.20063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman R. E. (1969). Development of the lateral musculature in the teleost, Brachydanio rerio: a fine structural study. Am. Anat. J. 125, 457–493 10.1002/aja.1001250406 [DOI] [PubMed] [Google Scholar]
- White R. M., Cech J., Ratanasirintrawoot S., Lin C. Y., Rahl P. B., Burke C. J., et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522 10.1038/nature09882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 10.1038/nature10886 [DOI] [PubMed] [Google Scholar]
- Willis M. S., Schisler J. C., Portbury A. L., Patterson C. (2009). Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc. Res. 81, 439–448 10.1093/cvr/cvn289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. J., Qadota H., Mains P. E., Benian G. M. (2012). UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of Caenorhabditis elegans. Mol. Biol. Cell 23, 2623–2634 10.1091/mbc.E12-01-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wang Z., Hu Y., Wu Y., Luo Z., Yang Z., et al. (2013). Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41, e141 10.1093/nar/gkt464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Tong X., Wang Z., Liu D., Pan R., Li Z., et al. (2013). TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10, 329–331 10.1038/nmeth.2374 [DOI] [PubMed] [Google Scholar]


