Abstract
Background
In the model single-cell C4 plant Bienertia sinuspersici, chloroplast- and nuclear-encoded photosynthetic enzymes, characteristically confined to either bundle sheath or mesophyll cells in Kranz-type C4 leaves, all occur together within individual leaf chlorenchyma cells. Intracellular separation of dimorphic chloroplasts and key enzymes within central and peripheral compartments allow for C4 carbon fixation analogous to NAD-malic enzyme (NAD-ME) Kranz type species. Several methods were used to investigate dimorphic chloroplast differentiation in B. sinuspersici.
Results
Confocal analysis revealed that Rubisco-containing chloroplasts in the central compartment chloroplasts (CCC) contained more photosystem II proteins than the peripheral compartment chloroplasts (PCC) which contain pyruvate,Pi dikinase (PPDK), a pattern analogous to the cell type-specific chloroplasts of many Kranz type NAD-ME species. Transient expression analysis using GFP fusion constructs containing various lengths of a B. sinuspersici Rubisco small subunit (RbcS) gene and the transit peptide of PPDK revealed that their import was not specific to either chloroplast type. Immunolocalization showed the rbcL-specific mRNA binding protein RLSB to be selectively localized to the CCC in B. sinuspersici, and to Rubisco-containing BS chloroplasts in the closely related Kranz species Suaeda taxifolia. Comparative fluorescence analyses were made using redox-sensitive and insensitive GFP forms, as well comparative staining using the peroxidase indicator 3,3-diaminobenzidine (DAB), which demonstrated differences in stromal redox potential, with the CCC having a more negative potential than the PCC.
Conclusions
Both CCC RLSB localization and the differential chloroplast redox state are suggested to have a role in post-transcriptional rbcL expression.
Keywords: Single-cell C4 photosynthesis, Bienertia sinuspersici, Dimorphic chloroplasts, Chloroplast differentiation
Background
C4 photosynthesis combines two distinct sets of carboxylation reactions that work as a biochemical CO2 pump, to increase the efficiency of CO2 fixation by ribulose 1,5 bisphosphate carboxylase oxygenase (Rubisco) [1-3]. The first set of C4 reactions begins with the initial generation of phosphoenolpyruvate (PEP) by pyruvate, Pi dikinase (PPDK). Then, PEP is used in the carboxylation action of phosphoenolpyruvate carboxylase (PEPC), which mediates the assimilation of atmospheric CO2 into C4 acids (malate or aspartate). The second set of reactions occurs when the C4 acids are transported to an internalized Rubisco-containing cell or compartment, where they are subsequently decarboxylated by either NADP malic enzyme (NADP-ME), NAD malic enzyme (NAD-ME) or PEP carboxykinase (PEPCK), depending on the C4 species. C4 photosynthesis is typically carried out using a dual-cell system known as Kranz anatomy, where the C4 acids are first produced in mesophyll (M) cells, and then transported to the bundle sheath (BS) cells for decarboxylation and re-fixation of CO2 by Rubisco. A primary goal of C4 photosynthesis research is to understand the process of dimorphic chloroplasts formation and the extent that the chloroplast differentiate from one another in order to support C4 biochemistry.
C4 differentiation, including the formation of morphologically and functionally dimorphic chloroplasts, occurs during leaf development in both monocot and dicot C4 species. The development of full C4 capacity progresses from early to mature developmental stages, as structural characteristics and expression patterns for genes encoding various photosynthetic enzymes diverge between the two cell types. Developmental patterns of chloroplast structural and functional differentiation among different C4 species show similarities as well as differences [4]. For example, in Amaranthus hypochondriacus (NAD-ME type eudicot) and Zea mays (NADP-ME type monocot) very early in leaf development Rubisco is initially present in both M and BS cell chloroplasts, in a non-C4 pattern, until developmental cues or light signals lead to its restricted accumulation to BS cells alone [5-7]. Complete biochemical and structural differentiation of M and BS cells and the characteristic dimorphic C4 chloroplasts is finalized during leaf ontogeny. In addition to differences between the two chloroplast types in Rubisco accumulation, a significant manifestation of this process is differentiation in grana development and the relative levels of components of photosystem I (PS I) versus photosystem II (PS II) in mature leaves [7-10]. The BS chloroplasts of NADP-ME species are deficient in grana stacks due to depleted nuclear encoded PS II components [11], whereas the BS chloroplast of NAD-ME species have more grana development and PS II content compared to M chloroplast [9,12]. While thylakoid differentiation is thought to be regulated mostly by energy requirements (ATP and NADPH) in M and BS cells to support the forms of C4, the underlying regulatory mechanism(s) responsible for C4-associated differentiation remains a very active area of research [2,13-16].
The control of cell-type specific differentiation in C4 species with Kranz anatomy has long been considered to be founded primarily on the opposing transcriptional activation/inactivation of select photosynthetic genes within the nuclei of the distinct M and BS cells. Thus, it was surprising when dimorphic chloroplasts performing full C4 photosynthesis were discovered within single chlorenchyma cells of some very unique C4 species (see [17]). Over 7,500 C4 species are currently known to exist [18], with Bienertia sinuspersici, in family Chenopodiaceae, being just one of four known terrestrial species that can perform single-cell C4[19,20]. Bienertia functions analogous to Kranz C4 species, in that its dimorphic chloroplasts work together to concentrate CO2 at the site of Rubisco; however, it accomplishes this by their spatial separation between two cytoplasmic domains within individual chlorenchyma cells. Biochemically, Bienertia is classified as an NAD-ME type C4, with decarboxylation of C4 acids in the C4 cycle occurring in mitochondria in a cytoplasmic domain known as the central compartment (CC) where the Rubisco-containing chloroplasts are also located [21,22]. The single-cell C4 system is thus unique in that there is only one nucleus for the transcription of genes encoding photosynthetic proteins that accumulate specifically within only one cellular compartment and, most notably, within only one of the two compartmentalized chloroplast types. Thus, post-transcriptional processes are, by necessity, required for the selective accumulation of these proteins to develop the dimorphic, compartmentalized chloroplasts which are required for C4 function in these plants.
Rubisco is a hetero-octomer, composed of an equal number of large subunits (rbcL) transcribed and translated in the chloroplast and small subunits (RbcS) that are transcribed in the nucleus, translated in the cytoplasm, and imported into the chloroplast [23]. As observed in some Kranz species, in the single-cell C4 species Suaeda aralocaspica and Bienertia cycloptera, Rubisco is initially present in both chloroplast types early in development [22,24]. There have been numerous studies exploring Rubisco assembly, which show that rbcL and RbcS subunits accumulate in equal amounts within the chloroplast stroma, and the loss of either peptide will cause a decrease in the other subunit peptide [25]. Recently, an RNA binding protein known as RLSB has been shown to correlate with BS specific localization of Rubisco in both monocot and dicot C4 plant species. RLSB is hypothesized to be necessary for rbcL mRNA maturation and translation in both C3 and C4 plants [26]. There are various ways that selective accumulation of Rubisco to the chloroplasts in the CC of Bienertia could be controlled; e.g. selective chloroplast targeting of nuclear encoded proteins such as the RbcS itself, a Rubisco-associated chaperonin such as Raf1 [27] or RLSB. Other mechanisms could include the selective transcription or translation of rbcL within the CCC, regulated assembly, selective degradation of necessary assembly chaperones, or some combination of these mechanisms.
In situ immunolocalization and western blots using isolated chloroplasts have demonstrated selective accumulation of Rubisco within the CCC of mature leaves of Bienertia[21,22]. These methods have also shown the selective compartmentalization of PPDK to the other chloroplast type within the peripheral compartment (PC). In addition to these selectively targeted proteins between these two chloroplasts, there are other nuclear-encoded proteins with a biochemical role in both chloroplast types that would need to be dual targeted. This group includes the enzyme pyrophosphatase [which is required in starch synthesis in the CCC and in the generation of phosphoenolpyruvate (PEP) from pyruvate in the PCC], and enzymes of the reductive phase of the C3 cycle (phosphoglycerate kinase and glyceraldehyde-P dehydrogenase) [21]. The goal of the current study was to investigate the dimorphic chloroplasts of Bienertia, with a central focus on mechanisms that may be responsible for the selective accumulation of Rubisco and PPDK, two enzymes that are specific to different chloroplast types located in separated domains. Specifically, we have investigated the possible selective targeting of RbcS to the CCC and PPDK to the PCC, through the use of constructs containing various lengths of the RbcS transcript fused to GFP, as well as the transit peptide of PPDK, by use of biolistic and protoplast transformation of Bienertia chlorenchyma cells. Cellular localization of the RLSB protein was determined as another possible control mechanism of rbcL synthesis. To assess the redox status as a potential control mechanism for each chloroplast type, fluorescence from a redox sensitive GFP (roGFP2) was measured, and an in vivo peroxidase activity and hydrogen peroxide (H2O2) stain was quantified. We discuss evidence that overlapping regulatory processes could act as determinants in the formation and function of dimorphic chloroplasts in this single cell C4 system.
Results
GFP expression analysis
Summary of GFP Constructs. The set of GFP fusion constructs were made by placing Bienertia c-DNA (except the AGPase construct [28]) on the N-terminus of the GFP protein (except the 3′UTR of RbcS). A list of the constructs used and results are summarized in Additional file 1: Table S1. Briefly, puc18 spGFP is a positive control lacking a targeting signal. PPDK constructs contained the first 180 or 273 protein-encoding nucleotides (PPDK-180 spGFP and PPDK 273 spGFP) while PPDK-CDS spGFP has the entire PPDK coding sequence (CDS). RbcS constructs contained the first 252 or 273 protein-encoding nucleotides (RbcS-252 spGFP and RbcS-273 spGFP) of the most abundant of three Bienertia RbcS transcripts [29] (R. Sharpe, unpublished), while RbcS-CDS spGFP has the entire RbcS CDS. Additional RbcS UTR sequence was added in various combinations to the RbcS constructs, with the RbcS-FL spGFP construct containing the entire Bienertia RbcS transcript. Alternative RbcS-FL spGFP constructs included using the super ubiquitin promoter (pSU) with an intron (pSU RbcS-FL spGFP) and re-placing spGFP protein with the roGFP2 protein (RbcS-FL roGFP2). A construct containing the entire CDS of the Bienertia RLSB c-DNA was fused in frame to the N-terminus to the GFP protein (RLSB CDS spGFP).
Biolistic Transformation. Initially biolistic transformation was used to test for expression and plastid targeting of GFP constructs in Bienertia leaves compared with onion epidermal cells (see results with biolistics in Additional files 2, 3, 4, 5). In onion, various constructs were expressed in epidermal cells, and in many cases import into pro-plastids was observed. In Bienertia most constructs were expressed in chlorenchyma cells; but, only rarely was import into chloroplasts observed (i.e. see evidence for import into both PCC and CCC with construct PPDK-180 spGFP and RbcS-273 spGFP, Additional files 3 and 4: Figure S2 and S3). Due to the limited occurrence of import in Bienertia with biolistics, to further evaluate expression and targeting using these constructs, protoplast transformation was used.
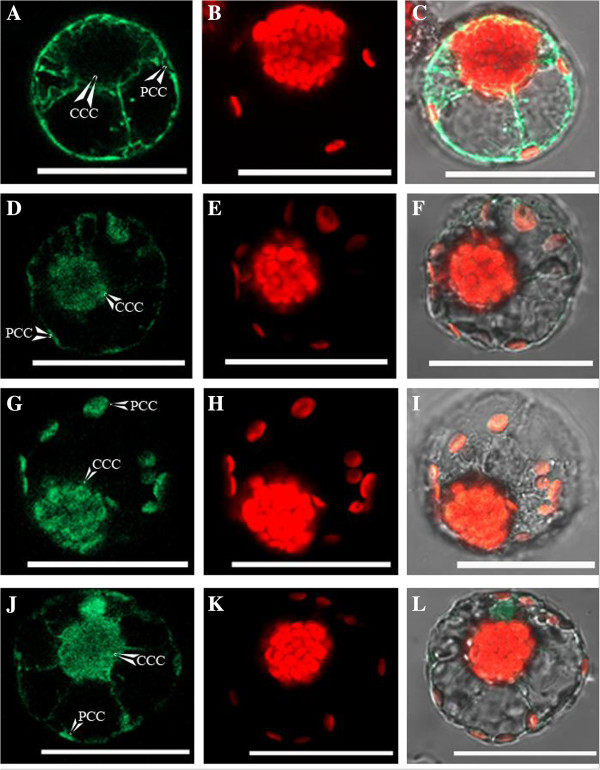
Plastid targeting of GFP constructs using protoplast transformation. A summary of the results with transient expression of constructs in Bienertia chlorenchyma protoplasts shows a consistent high co-occurrence of import with that which occurs in onion by biolistic treatment (Additional file 1: Table S1). The puc18-spGFP positive control showed GFP expression throughout the cytoplasm in Bienertia, with no GFP fluorescence observed within any of the chloroplasts (Figure 1, A-C), as also observed with the biolistic method with onion and Bienertia. The PPDK180-spGFP construct showed significant levels of GFP expression in both the PCC and CCC (Figure 1, D-F). Also, the RbcS273-spGFP construct produced clearly observable GFP expression in both the PCC and the CCC (Figure 1, G-I). The pSU-RbcS full length-spGFP construct showed GFP expression in both the PCC and CCC, with some GFP accumulation observed within the nucleus as well (Figure 1, J-L) (to some extent GFP translocation occurs into the nucleus [30]). Results for the RLSB-spGFP construct in Bienertia protoplasts were inconclusive, due to very low levels of GFP expression. Quantitative measurements are not shown for constructs that had similar GFP fluorescence from the PCC and CCC.
Figure 1.
Confocal images of GFP expression in Bienertia protoplasts. A - C is spGFP (positive control), D – F is PPDK180-spGFP, G – I is RbcS-273 spGFP, J – L is pSU RbcS-FL spGFP. Images A, D, G, and J are GFP emission. Images B, E, H, and K, are chlorophyll autofluorescence emission. Images C, F, I, and L are the merged images of GFP and chlorophyll autofluorescence emission. CCC = central compartment chloroplast, PCC = peripheral compartment chloroplast. Scale Bar = 50 μm.
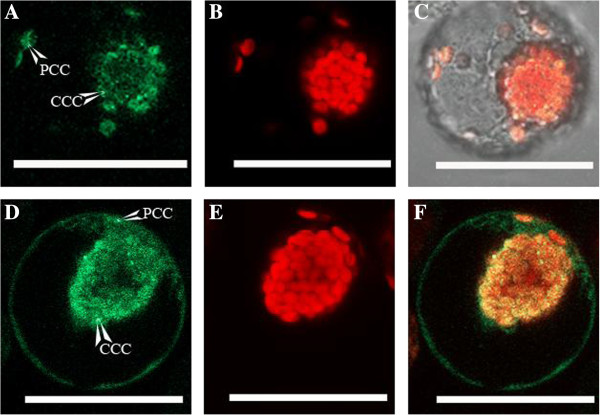
Because of the high sensitivity for the detection of GFP expression and targeting, as well as the lack of selectivity for import into either chloroplast type, the protoplast transformation method was used to determine the relative redox states of the dimorphic chloroplasts of Bienertia. For this purpose, protoplasts were transformed using the RbcS-FL construct linked with spGFP (redox insensitive) or roGFP2 (redox sensitive) reporter proteins (Figure 2). Representative data of confocal quantification of GFP fluorescence from the dimorphic chloroplasts of Bienertia protoplasts is shown in Additional file 6: Figure S5. Both constructs, which only differ in the redox sensitivity of their attached GFP protein, correctly target GFP to both chloroplast types (Figure 2, A and C). Comparatively analyzing GFP fluorescence between the two-chloroplast types indicted that fluorescence intensity from the spGFP protein was similar in both, while the roGFP2 protein produced a higher intensity of GFP fluorescence within the CCC than within the PCC. A summary of the confocal microscopy quantification of GFP fluorescence from transformed Bienertia protoplasts using Lambda mode, which quantifies fluoresence intensity from multiple locations, is presented in Table 1. The results show that the average CCC/PCC ratio of GFP fluorescence when using the redox insensitive spGFP protein was 1.028 ± .024, while the average ratio of GFP fluorescence between the two chloroplast types when using the redox-sensitive roGFP2 protein was 1.439 ± .005. The fluorescence of the roGFP2 protein, when excited with 488 nm, is known to decrease as the redox potential of its environment is decreased [31]. Attempts to determine an exact mid-point potential of the two chloroplast types were made, by making ratiometry measurements of fluorescence. However, the fragile nature of the protoplasts were not suitable for such measurments, due to the requirement for calibration of the system by additional washings of the protoplasts with reducing and oxidizing solutions.
Figure 2.
Confocal images of GFP expression in Bienertia protoplasts from the construct RbcS-FL spGFP (A-C) and the construct RbcS-FL roGFP2 (D-F). The roGFP2 protein is redox sensitive, while the spGFP protein is redox insensitive. Cells were scanned for a focal plane closest to the slide surface to maximize GFP fluorescence from each chloroplast type. Images A and D are GFP emission. Images B and E are chlorophyll autofluorescence. Images C and F are the merged images of GFP and chlorophyll autofluorescence emission. CCC = central compartment chloroplast, PCC = peripheral compartment chloroplast. Scale Bar = 50 μm.
Table 1.
The average CCC/PCC ratio of GFP fluorescence from the dimorphic chloroplast of transformed Bienertia protoplasts, using the constructs RbcS-FL spGFP and RbcS-FL roGFP2, where the roGFP2 protein is redox sensitive while the spGFP protein is redox insensitive
| Construct | # of quantified cells | Average CCC/PCC ratio of fluorescence |
|---|---|---|
| RbcS-FL spGFP |
31 |
1.028 +/− 0.024* |
| RbcS-FL roGFP2 | 28 | 1.439 +/− 0.005* |
Lambda mode on the Zeiss 510 confocal microscope was used to measure the intensity of GFP fluorescence. Fluorescence intensity values were obtained using an excitation wavelength of 488 nm, and detection of emission at a wavelength of 513 nm. (Additional file 6: Figure S5 shows a picture of the analysis). The ratio of GFP fluorescence intensity between the two chloroplast types was calculated by dividing the intensity of one central compartment chloroplast by the intensity of one peripheral compartment chloroplast. The ratio was averaged across the number of quantified cells. The results presented are from 7 biological replicates. The standard error from all measurments is shown. *Difference between the two constructs are statistically significant at p < 0.05, using a independent T-test.
Immunolocalization
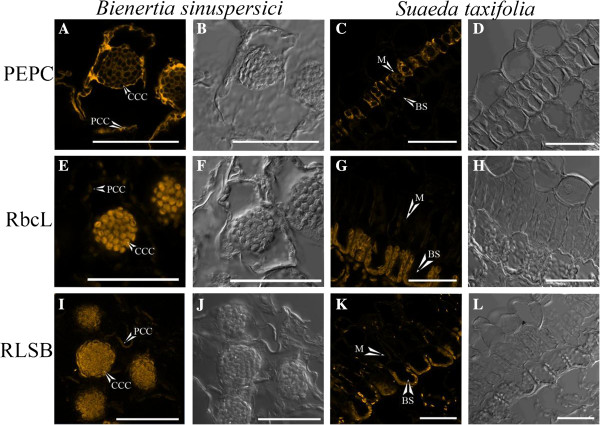
Immunolocalization was used to determine the cellular localization of the C4-associated proteins PEPC, rbcL, and the regulatory RLSB within Bienertia leaf chloroenchyma cells, and, for comparison, in the closely related Kranz-type species S. taxifolia (Figure 3). Fluorescence from an Alexa fluor 546-tagged secondary antibody, reacting with the primary antisera was detected using confocal microscopy. The analysis demonstrated that PEPC, as expected, was distributed throughout the cytoplasm in Bienertia chloroenchyma cells, and was selectively localized in M cells of S. taxifolia leaves (Figure 3, A-D). rbcL showed selective localization within Bienertia chloroplasts, with very high levels of intensity occurring only within the CCC. In comparison, the rbcL protein was highly specific to the BS chloroplasts of S. taxifolia (Figure 3, E-H), in the characteristic pattern for Kranz leaves. Most significantly, the regulatory RLSB protein, like the rbcL protein it is proposed to regulate, was selectively localized only within the CCC of Bienertia chlorenchyma cells. In agreement with previous findings [26], RLSB accumulation was observed to be highly specific to the BS chloroplasts in Kranz leaves of S. taxifolia (Figure 3, I-L).
Figure 3.
Immunolocalization of C4 proteins in single cell type Bienertia and Kranz type Suaeda taxifolia. Confocal microscopy detection of the Alexa fluor 546 tagged secondary antibody, reacting to the primary antibodies indicated, showing the location of phosphoenolpyruvate carboxylase (PEPC, A-D), rubisco large subunit (rbcL, E-H), and rubisco large subunit mRNA binding protein (RLSB, I-L) in Bienertia(A, B, E, F, I, and J) and Suaeda taxifolia(C, D, G, H, K, and L). Images A, C, E, G, I, and K are Alexa Fluro 546 detection. Images B, D, F, H, J, and L are bright field view of sections. CCC = central compartment chloroplast, PCC = peripheral compartment chloroplast, M = Mesophyll Cell, BS = Bundle Sheath Cell. Scale bar = 50 μm.
It should be noted that within the Bienertia chlorenchyma cells, both rbcL and RLSB proteins showed very slight levels of detection within the PCC, possibly due to very low levels of accumulation, or possibly background levels of reaction within these chloroplasts. Taken together, these findings clearly demonstrate strong specific localization of RLSB to the Rubisco-containing CCC of Bienertia chlorenchymeca cells, and the BS cell chloroplasts of a closely related but structurally distinct C4 species.
DAB staining to test for relative redox state of CCC and PCC in bienertia
The use of scanning electron microscopy (SEM) backscattering to quantify the precipitation of DAB particles in Bienertia tissues proved to be more reproducibly accurate than light microscope or transmission electron microscopy (TEM) images (images not shown). This can be attributed to the limited handling requirements for SEM sample preparation, as well as reducing variability due to sectioning and staining protocols required for light microscope and TEM. The backscattering mode on the SEM produces images that correspond tightly to the density of electron particles present. The more electrons present in an area the brighter the area will be in the image. In contrast, where there are few electrons (i.e., only water present, as in the vacuole space), electron density is low, and the area appears black. Various staining times, and light intensities were tested, and conditions that maintained cell morphology and optimized visualization/quantification of staining levels were used for analysis.
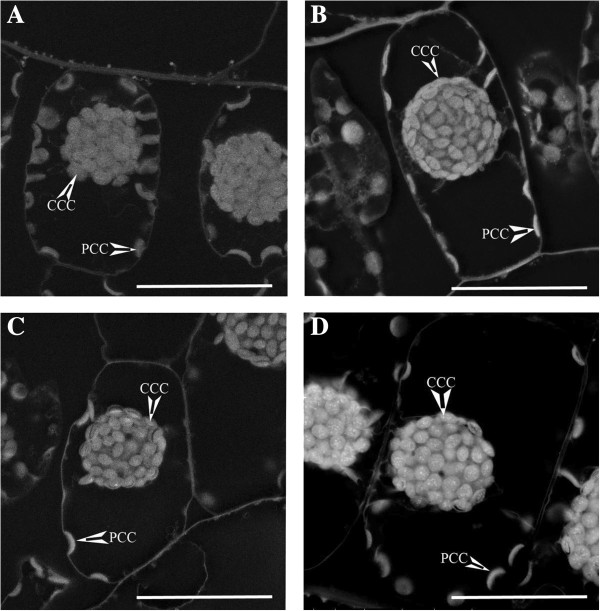
To probe for relative steady state levels of H2O2 generation in the dimorphic chloroplasts in the light, Bienertia leaves were excised and incubated under 100 PPFD (this low light intensity was used to avoid damaging the cells) in the presence of DAB (H2O2 stain) versus in the absence of DAB (control). Quantification of electron densities revealed that in the absence of DAB (negative control), the CCC was brighter in comparison to the PCC, with an average density ratio of 1.658 ± 0.128 (Figure 4-A, Table 2). When DAB was included in the staining reactions to detect H2O2 production, the PCC became brighter relative to the CCC, with the average density ratio dropping to 1.265 ± 0.097 (Figure 4-B, Table 2). Quantification and comparison of the CCC to PCC integrated density ratios showed that the density ratio of particles decreased when staining for H2O2, suggesting that more DAB was precipitated in the PCC than in the CCC. This is indicative of a higher level of H2O2 in the PCC relative to the CCC, which interacts with and precipitates DAB under these steady state conditions.
Figure 4.
Representative images from SEM backscattering detection of DAB stained Bienertia chlorenchyma cells (For quantification of results see Table 2). Image A is the control for H2O2 detection (no DAB was included). Image B is the stain for H2O2, where DAB was allowed to react with in vivo produced H2O2. Image C is the control for peroxidase activity (no H2O2 was added). Image D is the stain for peroxidase activity, where H2O2 is supplied in excess. CCC = central compartment chloroplast, PCC = peripheral compartment chloroplast. Scale Bar = 50 μm.
Table 2.
SEM backscattering quantification of Bienertia DAB staining, using Image J analysis to quantify the Integrated Density of the two chloroplast types
| Stain | Total average CCC/PCC density ratio |
|---|---|
| Control stain |
1.658 +/− 0.128* |
| H2O2 stain |
1.265 +/− 0.097* |
| Peroxidase control stain |
1.196 +/− 0.058* |
| Peroxidase stain | 1.965 +/− 0.239* |
The density ratio was calculated by dividing the average integrated density of 5–7 central compartment chloroplasts (CCC) by the average integrated density of an equal number of peripheral chloroplast (PCC) for each cell. The total average CCC/PCC density ratio was obtained by averaging the density ratio from two biological replicates (3 cells each) and two technical replicates. The standard error is shown for all 12 measurements. *Difference between the control and stain are statistically significant at p < 0.05, using a independent T-test.
To test for difference in the ability of the two chloroplasts to scavenge and reduce H2O2, a peroxidase activity stain was performed by adding H2O2 to the DAB staining solution. Excised leaves were placed in the DAB staining solution in the absence of H2O2 (peroxidase control where in vivo H202 is limiting), or in the DAB staining solution with 7 mM H2O2 (peroxidase stain) added. Analysis of the images indicated the two chloroplast types had a similar appearance with the control (H2O2 minus) peroxidase stain. In this control, the CCC was only slightly brighter than the PCC, with an average density ratio of 1.196 ± 0.058 (Figure 4-C, Table 2). When H2O2 was included in the stain to detect peroxidase activity, the CCC became significantly brighter than the PCC, so that the average density ratio rose to 1.965 ± 0.239 (Figure 4-D, Table 2). Upon closer observation of the peroxidase stain images, the CCC had more visible precipitation dots in comparison to PCC or the control images. Quantification and comparison of the CCC to PCC integrated density ratios showed that the density ratio increased significantly in this peroxidase activity stain. The results indicate that there is more peroxidase activity in the CCC relative to the PCC, under the steady state conditions of this assay.
Estimation of PSII Content between the two chloroplast types
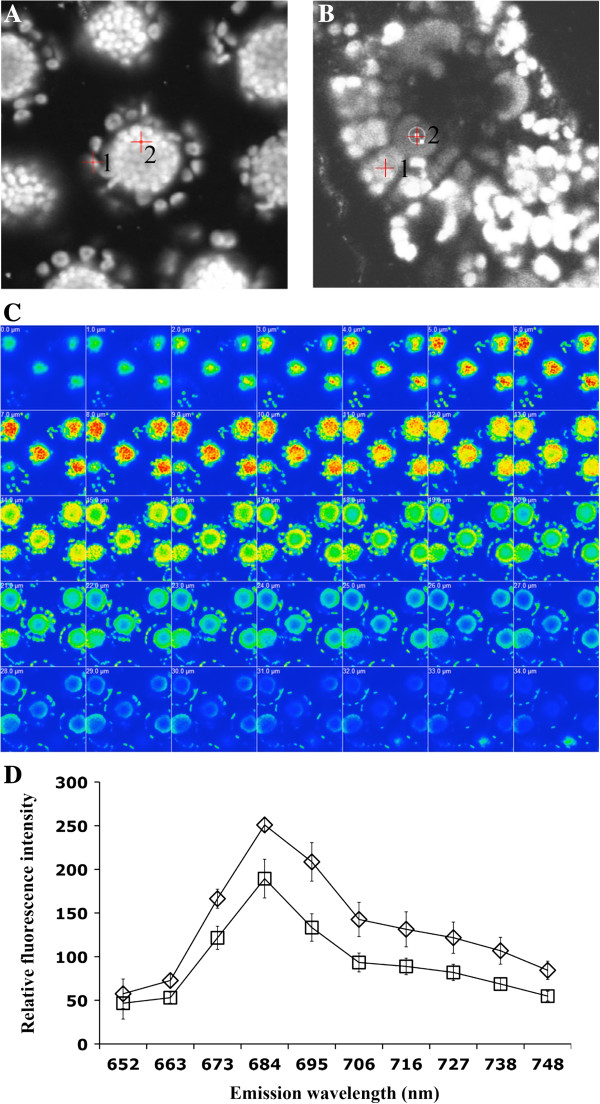
Quantification of chloroplast fluorescence emitted from PSII has been shown to be a rapid and efficient method for assessing differences in the PSII content in different chloroplast types of C4 species [32]. The application of this analysis to Bienertia protoplasts demonstrated that the CCC had a greater intensity of fluorescence intensity in comparison to the PCC (Figure 5-A), indicative of a higher PSII content in the CCC. As a comparative control, this same analysis was performed using a fresh cross-section of a Zea mays leaf, a Kranz type C4 species that is known to have low PSII content in BS chloroplasts (Figure 5-B). As expected, in this species the PSII-related fluorescence was greater from M (PSII containing) than BS (PSII reduced) chloroplasts.
Figure 5.
Estimation of PSII content in Bienertia chlorenchyma cells, using confocal microscopy. Cells were excited with 633 nm of light, and the brighter areas in images A, B, and C have greater fluorescence intensities, corresponding to more PSII content. A) Lambda mode image showing fluorescence from the two chloroplast types of Bienertia. Cursor 1 was placed on one peripheral chloroplast, while cursor two was placed on one central compartment chloroplast. B) Lambda mode image showing fluorescence from the two chloroplast types of Zea mays. Cursor 1 was placed on the mesophyll chloroplasts, while cursor two was placed on the bundle sheath. C) Heat map of a z-stack (serial images) series of a Bienertia protoplast at 1 μm slices, intensity represents the emission at 684 nm using 633 nm excitation. D) Quantification of the emission spectra of the two types of Bienertia chloroplasts; squares are peripheral compartment chloroplasts, and diamonds are central compartment chloroplasts. Error bars represent the standard deviation for 8 different protoplasts.
A heat map z-scan from a whole intact Bienertia protoplasts is shown in Figure 5-C, where the intensity of the heat signal (with red as the highest) corresponds to fluorescence at 684 nm. Within each protoplast frame, it can be seen that all of the detectable fluorescence emitted from the CCC has a much higher intensity than that emitted from the PCC. When averaged across 8 protoplasts, there is 1.3 times higher PSII associated fluorescence emitted from CCC compared to PCC (Figure 5-D).
Discussion
Photosystem II content in central and peripheral chloroplasts and its relevance to C4 biochemistry in Bienertia
Comparative confocal quantification is an established technique for determining the relative PS II content in M and BS cells of C4 species [32,33]. Applying this methodology to the dimorphic chloroplasts of Bienertia showed that the density of PSII is substantially higher in the CCC, which suggests higher capacity for linear electron flow than in the PCC. These relative amounts are comparable to prior quantitative analysis of dimorphic chloroplast in the single cell C4Bienertia cycloptera, which were shown to have a CCC granal index (the percentage of thylakoid membranes which are appressed) that is 1.5 times higher than the PCC [34]. This is typical for Kranz type species with NAD-ME biochemistry, with some species having up to 2-fold higher chloroplast granality in BS compared to M cells [9,33]. These results are consistent with the general assessment that there is an enrichment of PSII in BS chloroplasts relative to M chloroplasts in NAD-ME type species.
Chloroplast ultrastructural differentiation observed at the mature cell stage of Bienertia could be the result of a progressive establishment of C4 biochemistry during leaf ontology, since early in development there is little difference in the thylakoid membranes of the two chloroplasts [22]. This is true for other C4 species, where differences in the granal index of different chloroplast types are observed only later in development [35]. In mature leaves of Kranz-type NAD-ME species the only energy requirements in the C4 cycle is 2 ATP per CO2 delivered to the BS cells. For single cell C4 species such as Bienertia, this requirement for ATP occurs in the PCC to support the conversion of pyruvate to PEP. Thus there is a greater energy requirement in the CCC (as in the BS chloroplasts of NAD-ME Kranz species), where NADPH needs to be generated by PSII dependent electron transport to support the C3 cycle [4]. As C4 leaf development progresses, aspects of C4 biochemistry between the PCC and the CCC diverge, with the demand for NADPH decreasing in the PCC and increasing in the CCC. In mature leaf cells, reduced NADPH utilization in the PCC would likely cause an acceptor limitation for electrons derived from PSII activity, resulting in inactivation of PSII by generation of singlet oxygen, and a corresponding increase in cyclic electron flow for ATP generation by PSI [36,37]. This type of biochemical regulation of PSII content has also been demonstrated in transgenic rice (a C3 plant) by expressing Zea mays NADP-ME in the chloroplasts that resulted in reduced development of grana and PSII activity. This was suggested to be caused by increased uptake and decarboxylation of malate by NADP-ME in the chloroplasts which generates reductive power leading to reduced need for photosynthetic production of NADPH via PSII [38]. These studies, together with findings presented here, support a model in which the basic requirement for energy production, in itself, could be a determinant for the regulation of PS I and PS II development. Such dynamic regulation could ultimately balance the production/accumulation of the two photosystems according to the needs of each chloroplast type.
Mechanisms for selective chloroplast accumulation of nuclear encoded proteins
The unique single-cell C4 system of Bienertia differs significantly from the two-cell system found in the more typical Kranz species. By necessity this system is primarily regulated by post-transcriptional control processes that achieve selective accumulation of enzymes in the dimorphic chloroplasts. Several hypotheses have been recently proposed for differential protein localization in this system, including selective chloroplast protein import, mRNA trafficking to specific domains for translation, and/or the selective proteolysis after protein import [39].
Analysis of the various RbcS and PPDK GFP fusion constructs were used in this study to test for selective chloroplast import. The results showed that similar levels of GFP uptake occurred in both chloroplasts types in Bienertia chlorenchyma cells (Figure 1 and Additional files 3 and 4). A similar observation was made with RbcS in an earlier study, where an undefined RbcS sequence was shown to direct GFP import into both chloroplast types within Bienertia protoplasts [40]. Taken together, the in vivo biolistic and protoplast analysis of this current study clearly demonstrate that the transit peptide of PPDK or various lengths of the most abundant RbcS proteins, in themselves, were capable of directing at least some level of GFP fusion uptake, depending on the plant system used. However, the protein expressed from these constructs showed no evidence of selectivity for either Bienertia chloroplast type. The lack of selectivity in GFP targeting could also suggest that additional sequence information that was not included in the constructs might be needed for selective chloroplast targeting. This could include additional mRNA sequences (introns or more UTR), alternative gene family members in the case of RbcS, or differential placement of the GFP CDS. There could also be experimental effects, such as a difference in recognition and import of native peptides versus the peptide fragments, or regulation in intact leaves that becomes disrupted in protoplasts. For example, selective import/targeting could be disrupted during protoplast isolation and overnight incubation. However, observations by light microscopy showed the two cytoplasmic domains were maintained (which is dependent on integrity of the cytoskeleton [41]).
One potential mechanism for selective targeting of proteins within Bienertia chloroenchyma cells is the trafficking of mRNA facilitated by mRNA-binding protein (RBP) complexes. While cytoplasmic mRNAs typically associate with RBPs to control their stability or initiate and maintain translation on free ribosomes [42], there are some RBPs that can move selected mRNAs along the cytoskeleton to destinations within specific subcellular domains. As an example, some RBPs in plant cells can mediate trafficking to defined regions of the endoplasmic reticulum (ER). The most studied example is the OsTudor-staphylococcal nuclease (SN) RBP that facilitates the movement of prolamine mRNA along actin filaments to the cortical ER in rice endosperm [43]. However, to date there have been no reports of RBPs responsible for localizing nuclear-encoded photosynthetic transcripts, such as RbcS or C3 cycle enzymes, to specific domains of a photosynthetic cell (i.e. closer to the chloroplast for more efficient chloroplast targeting).
At least some regions necessary for RbcS regulation occur in the mRNA UTRs. In rice leaves, it was shown that for mRNA turnover to occur, there needs to be both the 5′ and 3′ UTR of RbcS present [44]. Similar results have been found in the C4 plants Zea mays and Cleome gynandra, where the RbcS UTRs or portions upstream of the gene (1.0 kb of ZmRbcs & 3.8 kb of CgRbcS gene region) were sufficient to confer BS specific RbcS or β-glucuronidase accumulation [45-47]. In themselves, the 5′ and 3′ UTRs of a heterologous RbcS mRNA from Amaranth, constitutively expressed in F. bidentis leaves, were sufficient to confer partial BS cell specificity of a β-glucuronidase fusion [45]. Taken together, these results imply that in the dual-cell Kranz system, interaction of UTRs with specific RBP(s) might enhance translation/stability in the BS cells, or perhaps decrease stability/translation in M cells. Further experimentation will be needed to address whether RbcS regulation in the cytoplasm is analogous to rbcL regulation in the chloroplasts, i.e. whether an RBP such as the chloroplastic RLSB [26] might be capable of interacting with and mediating RbcS transcript stability/translation in the cytoplasm. While this seems like a plausible mechanism, no RbcS mRNA interacting proteins of any type have been identified. In this study, the addition of the pSU intron upstream of the RbcS 5′UTR, along with the pSU plant promoter did not change the results of non-selective GFP fluorescence from the dimorphic chloroplasts (Figure 1). Thus, even when using a plant promoter with upstream RNA sequences and an intron, there was no evidence for transcript-mediated selectivity for single-cell type compartmentalization in vivo.
In agreement with our findings, recent analyses on the translocon of the outer envelope membrane of chloroplasts (TOC) of Bienertia using various chloroplast targeted TOC-GFP constructs and BsTOC159, BsTOC132, and BsTOC34 antibodies showed that both chloroplast types import and accumulate nuclear encoded TOC proteins non-selectively similarly to other plants [40,48]. The current understanding of stromal and TOC protein import suggests that the main role of cytosolic targeting factors/chaperones in chloroplast localization is to increase targeting efficiency and uptake rate by maintaining peptide recognition competency [49,50]. The only factor that has been determined to be necessary for the proper sorting to the stroma is a transit peptide, which interacts with the stromal HSP70 chaperone and the outer envelope membrane [49,51,52]. One interesting aspect of chloroplast protein import, is the regulation that has been shown to occur by the redox regulated Tic62 protein, which changes its location based on the stroma NADP+/NADPH ratio [53,54]. There have been no previous reports for Kranz C4 species that regulation occurs at the chloroplast import level.
In summary, the findings presented here, together with those of previous studies, have revealed no evidence for the chloroplast import machinery, selective mRNA trafficking, or compartment-selective activation/translation of nuclear-encoded photosynthetic mRNAs, having a direct role in selective protein accumulation for the dimorphic chloroplasts in Bienertia. Therefore, C4 regulation in the single-cell system must involve additional regulatory processes, acting either independently or in synergy with one or more of the mechanisms described above.
Rubisco synthesis, assembly, and relevance to selective localization in Bienertia dimorphic chloroplasts
Post-transcriptional regulation (translation and stability) is the primary level at which plastid-encoded protein such as rbcL are regulated. Similar to nuclear-encoded transcripts, the UTRs of plastid-encoded mRNAs play a key role in their regulation. Many studies have demonstrated that such regulation of plastidic RNA metabolism occurs through interactions with groups of nuclear-encoded chloroplast RBPs. These proteins fall into several classes, with molecular masses ranging from 38–60 kDa [55-58]. Some of these have been shown to assemble on the 5′UTR of psbA, psbC, and rbcL, and other plastidic transcripts [56,57]. For many of these proteins, their production, binding, or activity are often determined by light- or redox potential, such that binding to the 5′ UTR under activation conditions might enhance the mRNA′s translation, processing, or stability [58,59].
In the case of rbcL mRNA, it has been shown that the rbcL protein itself has an N-terminal RNA binding domain that could have a role in mediating its own translational arrest by binding its own 5′ UTR [60]. To date, MRL1 and RSLB are the only two nuclear-encoded proteins to be directly implicated in the regulation of rbcL mRNA metabolism through interaction with its 5′ regions [26,61]. The number of chloroplast regulatory factors required for the correct assembly of Rubisco continues to increase, with RLSB and RAF 1 & 2 being the most recent additions [26,27]. Both BSD2 and RAF1 have been identified as rbcL assembly factors that are necessary for the proper assembly of Rubisco within the BS chloroplasts of C4 plants [27,62]. In Zea mays, RAF1 is specific to BS chloroplasts, whereas BSD2 is localized in both M and BS chloroplasts, indicting different regulatory roles for these functionally-related proteins. Thus, the selective accumulation of the Rubisco holoenzyme in the CCC of Bienertia could involve a compartmentally-selective increase in either rbcL mRNA binding proteins or rbcL assembly chaperones. These mechanisms are not exclusive, and both could work together to achieve differential Rubisco accumulation in mature Bienertia leaf cells.
The results show that RLSB is highly specific to the CCC in Bienertia cells, co-localizing with rbcL within this same chloroplast type, which is clearly distinct from the cytosolic localization of PEPC (Figure 3). The cellular localization shown for rbcL and PEPC is the same as previously reported for Bienertia[22,34]. The co-localization of RLSB to the Rubisco-containing chloroplasts provides evidence for a regulatory role in the selective compartmentalized synthesis of rbcL in this single-cell system. As a comparison, RLSB was also found to be specifically localize to the rbcL-containing BS chloroplasts of the related Kranz type S. taxifolia, in agreement with its BS-specific localization and proposed regulatory role in the Kranz C4 species Flaveria bidentis, Zea mays and Setaria viridis[26]. Selective localization of RLSB to the CCC in Bienertia suggests that, as in Zea mays, this protein may function in the post-transcriptional regulation of rbcL synthesis, binding to rbcL mRNA to enhance its translation/stability specifically within these chloroplasts.
As with other C4 proteins in this unique single-cell system, mechanisms responsible for the selective accumulation of RLSB within the CCC are as yet unknown. Possibilities include selective import to the CCC, or possibly selective degradation of RLSB in the PCC if it was unable to bind to its target rbcL mRNA. Attempts to determine if RLSB could be selectively targeted to the CCC when fused to GFP were inconclusive using Bienertia protoplast transformation, due to the low level of GFP expression from this construct. Some Chlamydomonas reinhardtii proteins responsible for rbcL translation have been shown to dissociate from the rbcL 5′ UTR upon increasing concentrations of oxidized glutathione; these are subsequently replaced through binding of the rbcL protein to its own mRNA [60]. A similar processes was suggested to occur in the higher plant Populus deltoides[63]. If such self-regulation were to occur in the Bienertia PCC, then it might be expected that a basal level of rbcL protein would always be present, as a way to prevent its own translation. In fact, low levels of rbcL were detected in the PCC by immunolocalization, although it is uncertain whether this is free large subunit, the Rubisco holoenzyme, or possibly low levels of background due to non-specific interactions of the primary antibody (Figure 3, N. Koteyeva and E. Voznesenskaya, unpublished observations).
In Nicotiana tabacum and Zea mays, it was previously shown that in the absence of RbcS, rbcL is subject to translational repression, through an interaction of unassembled rbcL with its encoding rbcL transcript [control by epistasy (CES)], a posttranslational regulation mechanism [25,64]. If CES exists in C4 plants, by transcriptionally limiting the availability of RbcS for Rubisco assembly (perhaps in Bienertia through selective chloroplast targeting), than translation of rbcL would be repressed, thereby impeding Rubisco assembly. However, GFP fusion constructs possessing different lengths of an abundant RbcS transcript did not support the selective transport aspect of this hypothesis. Fusing GFP to the C-terminal of the RbcS transit peptide, the RbcS CDS, or even the entire RbcS transcript (with the RbcS 3′UTR sequence being attached to the C-terminus of GFP), indicated that the RbcS protein can be imported into both chloroplast types, with no import selectivity identified (Figure 1). These results are in agreement with a recent analysis in Zea mays, where ectopic expression of RbcS in the M cells, alone or in combination with the expression of a nuclear-encoded version of an rbcL peptide targeted to chloroplasts, did not lead to significant accumulation of Rubisco in M cells [64]. Taken together these results indicate that selective RbcS targeting, and an associated inability to assemble Rubisco within one chloroplast type, are not in themselves responsible for the selective compartmentalization of Rubisco in Bienertia leaves.
Changes to the stromal redox state within the chloroplasts have been shown to affect rbcL translation as well as Rubisco enzymatic function [65,66]. Synthesis of rbcL has also been found to correlate with changes in ROS production, detoxification, and the ratio of GSH/GSSG within the chloroplasts [67,68]. The intact Rubisco enzyme itself is known to be susceptible to H2O2. Low concentrations lead to enzyme inactivation; increasing concentrations can cause unfolding and the loss of secondary and tertiary structure, ultimately leading to increased protease susceptibility [65,69]. Thus, another hypothesis for differential Rubisco accumulation in the two chloroplasts types is based on stromal redox conditions. This would occur if the CCC had a higher GSH/GSSG ratio compared to the PCC, and the CCC had a lower H2O2 concentration and greater detoxification capabilities. A difference in the redox state of the stroma is supported by the observation that roGFP2, which most directly reflects the ratio of GSH/GSSG, has altered fluorescence in the two Bienertia chloroplast types. Results with DAB staining also suggest that the PCC has a higher in vivo H2O2 concentration, with the CCC having more peroxidase activity. The level of H2O2 in the chloroplast and the redox state of GSH/GSSG depends on a number of factors including the capacity of the chloroplast for linear electron flow, the demand of NADPH for carbon assimilation, the extent of the Mehler reaction (electron transfer from PSI to O2 leading to H202 synthesis), and the capacity to scavenge H2O2 using antioxidants ascorbate and glutathione.
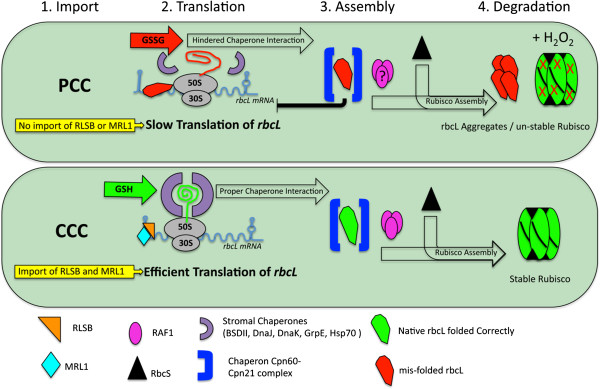
In this study, mechanisms responsible for selective Rubisco accumulation in forming dimorphic chloroplasts of mature Bienertia cells were investigated, a process which is necessary to support C4 biochemistry. Development of Bienertia leaves occurs acropetally. During development of the two cytoplasmic domains in young Bienertia chlorenchyma, some Rubisco is initially present in PCC as well as CCC, while mature chlorenchyma cells have strong selective accumulation of Rubisco only in the CCC [22]. These observations coincide with younger chlorenchyma having rbcL transcripts in both chloroplasts, while in mature chlorenchyma the transcripts are selectively expressed in the CCC (N. Koteyeva and E. Voznesenskaya, unpublished results). Findings from this study support a model in which the posttranscriptional regulatory activity of RLSB, together with variations in the plastid redox state, could function synergistically to activate/stabilize rbcL mRNA along with Rubisco in the CCC. The opposite effects would occur in the PCC, where repression/degradation of rbcL mRNA and Rubisco destabilization would prevent its accumulation (see details in the model in Figure 6). In Bienertia, control of rbcL translation may occur in coordination with increased transcript stability, possibly mediated by RLSB, as proposed for Zea mays[26]. In this case during Bienertia cell maturation, RLSB abundance could become progressively more chloroplast specific, so that rbcL mRNA and its encoded protein both become more abundant in the CCC, and less abundant in the PCC, in coordination with changes in photosynthetic electron transport and redox status. Under altered redox conditions in the PCC, rbcL may bind to rbcL, and prevent the binding of stablizing RBPs (like MRL1 and RLSB) leading to decreased rbcL transcripts. Additionally, elevated H2O2 concentrations in the PCC, could interfere with Rubisco assembly or cause degradation leading to less Rubisco.
Figure 6.
Regulatory hypotheses for how Rubisco becomes selectively localized to the central compartment chloroplasts (CCC) of Bienertia chlorenchyma cells. Regulatory point one, (yellow boxes) is the selective targeting or import of rbcL mRNA binding proteins (RBP) such as RLSB, or other proteins necessary for Rubisco assembly. Regulatory point two, is the translation of rbcL, where in the CCC under reduced conditions (GSH, green box) rbcL RBPs would increase translation of rbcL, while in the peripheral compartment chloroplasts (PCC) under elevated oxidized glutathione conditions (GSSG, red box), rbcL RBPs would not bind to rbcL, due to incorrectly folded rbcL binding to rbcL slowing translation. Regulatory point three, is the interaction of rbcL with stromal chaperonins and assembly of Rubisco, which in the PCC could be impeded due to a lack of chaperonins like RAF1 or the redox state of the chloroplast. Regulatory point four, is the stability or degradation of Rubisco which might be increased in the PCC due to elevated H2O2 concentrations. Figure adapted from [23,27,66].
Conclusions
The evidence presented here provides background that allows us to construct an initial model of selective Rubisco compartmentalization within single chlorenchyma cells of the single-cell C4 plant Bienertia (Figure 6). This model accounts for the fact that differential transcription of nuclear-encoded genes cannot be responsible for the selective compartmentalization of photosynthetic or regulatory proteins within a single cell. This model proposes that the rbcL post-transcriptional regulator factor RLSB (and possibly others), and differential redox status of the dimorphic chloroplasts, work together to restrict Rubisco accumulation to only one of the two chloroplast types of Bienertia chlorenchyma cells. This model is in concurrence with findings from many studies [27,70,71] demonstrating that selective accumulation of Rubisco in C4 systems is accomplished primarily by post-transcriptional mechanisms. Control of gene expression at this level is an absolute requirement to achieve the differential Rubisco accumulation patterns that characterizes C4 photosynthesis in Bienertia.
Methods
Plant material
Bienertia sinuspersici (hereafter referred to as Bienertia) plants were grown in controlled environmental chambers (Econair GC-16; Bio Chambers). Seedlings or vegetative cuttings were started under low light [100 PPFD (μmol quanta m-2 s-1) and temperature conditions with a day/night temperature of 25/22˚C and a photoperiod of 14/10 h]. Plants were moved to high light and temperature conditions (1000 PPFD, with a day/night temperature of 35/25˚C and a photoperiod of 14/10 h) once several branches were present. Mature leaves (~ 3 cm long) from 2- to 6- month old plants were routinely used for biolistic and protoplast transformation.
Construct assembly
Fusion protein constructs were made by subcloning DNA fragments of interest into the 35S:puc18-spGFP6 vector using restriction enzymes BamHI/NheI for N-terminal GFP addition, and restriction enzymes XbaI/SpeI for the C-terminal addition of the RbcS 3′ un-translated region (UTR) (M. Tegeder, unpublished data) (New England BioLabs, Ipswich, MA). DNA fragments of RbcS, and BADH were obtained from a cDNA library prepared using CloneTech SMARTer PCR cDNA synthesis kit according to manufacture protocols (Mountain View, CA). The PPDK fragments were amplified from a previous c-DNA library preparation [29]. DNA fragments were amplified using gene specific primers with restriction enzyme cut sites added for CDS fragments, or with the smart oligo primer, with one gene specific primer (Additional file 1: Table S2). The PCR fragments were digested and ligated into the puc18 vector using T4 DNA ligase (New England BioLabs, Ipswich, MA). Plasmid DNA of all constructs was purified using Zymo Research Plasmid DNA miniprep kits (Irvine, CA) and verified by sequencing (WSU Sequencing Core) before transformation (sequence of DNA fragments used in constructs are shown in Additional file 7: Figure S6). The CDS for roGFP2 was inserted into the puc18 vector, by PCR amplifying the roGFP2 fragment from the pRSETB vector [31], using primers that added the restriction enzyme cut sites NheI and XbaI to the PCR fragment (Additional file 1: Table S2). The PCR fragment and puc18 vector were digested with NheI and XbaI, ligated together using T4 DNA ligase, and verified by sequencing. The addition of the plant super ubiquitin promoter (pSU) and pSU intron, to the full-length RbcS-GFP fragment was done by first cutting out the full-length RbcS-GFP fragment using the enzymes BamHI and HindIII and ligated into Tegeder vector 656 (M. Tegeder, unpublished data). The pSU promoter and intron were subsequently added to vector 656 using the enzymes SacII and BamHI from Tegeder vector 655 (M. Tegeder, unpublished data). The construct with the transit peptide of AGPase fused to GFP was from [28], to test targeting of the large subunit of AGPase.
Biolistic transformation
Constructs were introduced into onion epidermal cells, spinach leaves, and Bienertia chlorenchyma cells by particle bombardment according to [72] and the manufacture’s protocol (Bio-Rad). For Bienertia bombardment, the epidermis of the leaf was first gently removed using forceps to grab the tip of the leaf and a small plastic pestle to gently press out chlorenchyma and internal leaf cells, resulting in an epidermis-less leaf. Cells were bombarded with 1 μm plasmid coated gold micro-carriers, using 1100 p.s.i. rupture discs at a distance of 6 cm from the stop screen. After bombardment epidermal free leaves were incubated overnight in a buffer (MES-NaOH, pH 5.8) that was osmotically adjusted with glycine betaine to match the osmolality of the leaf (determined with a 5500 Vapor Pressure Osmometer, Wescor). Bombarded onion epidermal cells, and spinach leaves were incubated overnight at room temperature on water moistened filter paper.
Protoplast isolation
Intact protoplasts were obtained from mechanically isolated chlorenchyma cells, using the protocol similar to [21] and [40]. Briefly, the leaf osmolality of plant material was measured prior to isolation and subsequently all buffers were osmotically adjusted to the measured value. Approximately 150 mature leaves (longer than 1 cm) were removed using a razor blade, early in the photoperiod. Chlorenchyma cells were gently pressed out using a motor and pestle into 8 ml of protoplast buffer (PB) (5 mM MES-NaOH, pH 5.8, 10 mM CaCl2, 0.7 M Sucrose, 1% Dextran, and matching glycine betaine concentration). The isolated cells were filtered through a nylon mesh with 1-mm pore size to remove large leaf particles. Cells were washed twice with PB by removing cells from the top of the 15 mL tube after centrifugation (5 min, 3.5 g in a Damon IEC HN-SII centrifuge). 1 mL of cells was mixed with 1 mL of digestion buffer (PB plus 2% [w/v] Sumizyme C [Shin-Nihon Chemical], 0.25% [w/v] Macerase [Calbiochem], and matching glycine betaine concentration) in a 2 mL round-bottom centrifuge tubes. Cells were incubated for 40 minutes at 35˚C on a Gyromax 737 orbital incubator shaker (Amerex Instruments) at 65 rpm with illumination from a 100-W light bulb to obtain protoplasts. The enzyme/cell mixture was centrifuged at 100 g for 2 min in a benchtop centrifuge and the floating layer of protoplasts were collected and washed twice with PB.
Protoplast transformation
The initial procedure used was developed based on [73], and subsequently modified using the protocol of [40]. Briefly, isolated protoplasts were incubated on ice for 30 min in W5 solution (5 mM MES-NaOH, pH 5.8, 154 mM NaCl, 125 mM CaCl2 and 5 mM KCl). Approximately 1.5 × 104 protoplasts were resuspended in chilled-protoplast buffer (5 mM MES-NaOH pH 5.8, and matching glycine betaine concentration) and mixed with 5 μg of plasmid DNA. The protoplast/DNA mixture was gently mixed with PEG (Sigma-Aldrich, St. Louis, MO, USA) solution (40% [w/v] in PB) to initiate a 5 min transfection at room temperature. Transfected protoplasts were mixed with 5 volumes of protoplast buffer, centrifuged for 1 min at 15 g, resuspended in 100 μl of protoplast buffer and subsequently cultured overnight at 25°C with a light intensity of 25 μmol m-2 s-1.
Protein immunolocalization in situ
Tissue was prepared and analyzed according to the procedures of [35]. Leaf samples for Bienertia and S. eltonica were fixed in FAA (50% ethanol, 5% glacial acetic acid, 10% formalin) fixative overnight at room temperature. Samples were dehydrated using increasing concentrations of ethanol, washed using CitriSolv, and embedded in Paraplast Plus. The paraffin-embedded samples were sectioned (thickness approximately 7.5 μm) using a rotary microtome, mounted on a poly-L-lysine-coated slides (Electron Microscopy Sciences, PA), and dried overnight at room temperature. After deparaffinization by CitriSolv, the sections were rehydrated through an ethanol series, and rinsed with H2O twice. Slides were washed twice with PBS buffer, and subsequently incubated with goat serum for 30 min at room temperature. Slides were washed twice with PBS buffer containing 0.1% BSA. Samples were incubated with diluted antibody for either, 2 h for PEPC and rbcL, or overnight for RLSB, in PBS buffer containing 0.1% BSA (PEPC was diluted 1:500; source [6], rbcL was diluted 1:1000; source [74], RLSB was diluted 1:250; source [26]. Slides were washed three times with PBS buffer containing 0.1% BSA, and incubated for 1 hour with the Alexafluor 546 conjugated secondary antibody (Invitrogen, Grand Island, NY), diluted 1:400 in PBS buffer containing 0.1% BSA. Samples were washed three times in PBS buffer containing 0.1% BSA, and subsequently rinsed three times in PBS buffer. Slides were mounted with vectashield (Vector Laboratories, CA), and sealed with a cover slip and nail polish, prior to visualization on the Ziess 710LSM confocal microscope (Imaging Facility, Dept. of Biological Sciences, University at Buffalo).
Confocal microscopy
Quantification of PSII Fluorescence – This procedure was adapted from [33]. Microscopy of fresh protoplasts was carried out using a Laser-Scanning-Microscope LSM 510 invert (Carl Zeiss, Oberkochen, Germany). The excitation wavelength for chlorophyll autofluorescence was 544 nm from a HeNe laser and the emitted wavelength captured for the images was long-band pass filter at 654 nm (Carl Zeiss). Images were obtained by averaging 8 measurements in frame mode. In this mode, the frame is scanned repeatedly and the signal is averaged. To quantify the relative intensities of chlorophyll fluorescence on individual chloroplasts from the central and peripheral compartments, the lambda mode was used.
GFP expression analysis - Approximately 100 μl of protoplasts were imaged on cover glass slides using a Zeiss 510 LSM. Serial Z-stack images were acquired at 1 μm intervals using a 63× water-immersion lens at a digital resolution of 1024 × 1024. The excitation wavelength for chlorophyll autofluorescence was 544 nm and the emitted wavelength captured for the images was long-band pass filter at 654 nm. GFP was excited using a wavelength of 488 nm and the emission detected at a band path of 505–530 nm. All images were further processed and composed using Adobe Photoshop CS5 (Adobe Systems Incorporated, Seattle, USA). All experiments were repeated at least three independent times with similar results. For qualitative analysis of expression, lambda mode was used to assess the intensity of GFP fluorescence from each transformed Bienertia protoplast. Fluorescence intensity values were obtained using an excitation wavelength of 488 nm, and detection of emission at a wavelength of 513 nm. In each cell analyzed, the ratio of GFP fluorescence intensity (CCC/PCC) from measurements on individual chloroplasts was determined. The ratios were averaged across the number of quantified cells, and the results presented are from 7 biological replicates. Statistical analysis was performed using an independent t-test using the Stastica 7 software (StatSoft Inc., Tulsa, OK, USA).
DAB staining
Mature leaves were cut into 3–4 mm sections using a super sharp razor blade. Leaf sections were placed into a glass vial for staining. Four stains (H2O2 control, H2O2 detection, Peroxidase control and Peroxidase detection) were performed using Sigma FAST 3, 3′ – Diaminobenzidine (DAB) Tablets (Sigma Aldrich, St. Louis MO) carried out at 25°C. Tablets were dissolved in water that contained a glycine betaine concentration that osmotically matched the leaves (DAB 0.7 mg ml-1, 7 mM H2O2, 60 mM Tris buffer pH = 7.5). For detection of the generation of H2O2 the tissue was incubated in the presence of DAB under 100 PPFD for 4 h (the control was without DAB). For peroxidase detection, leaf sections were incubated in the presence of DAB for 45 min with addition of 7 mM H2O2 under 10 PPFD (control was without H2O2). After staining leaves were fixed following [75]. After imbedding the plane of the cut leaf surface was sectioned down to using a Reichert Ultracut R ultramicrotome (Reichert-Jung GmbH, Heidelberg, Germany). The sample was super-glued (The Original Super Glue) onto an aluminum specimen mount (TED Pella) and imaged using the back-scattering detector under high vacuum mode at 30 kV accelerating voltage using a Quanta 200 F environmental field emission gun scanning electron microscope (FEI Company; Field Emission Instruments). Image quantification was done using Image J software on an equal number of PC and CC, typically 5–7 chloroplasts per cell depending on the number of PC present. A total of 3 cells were quantified per replicate, with two technical replicates, and two biological replicates being quantified. Statistical analysis was done using an independent t-test using the Stastica 7 software (StatSoft Inc., Tulsa, OK, USA).
Abbreviations
AMP: Adenosine monophosphate; ATP: Adenosine-5'-triphosphate; BS: Bundle Sheath cell; CC: Central compartment; CCC: Central compartment chloroplast; CDS: Coding sequence; CES: Control by the epistasy of synthesis; DAB: 3,3-Diaminobenzidine; M: Mesophyll cell; NAD-ME: NAD-malic enzyme; PB: Protoplast buffer; PC: Peripheral compartment; PCC: Peripheral compartment chloroplast; PEP: phosphoenolpyruvate; PEPC: Phosphoenolypyruvate carboxylase; Pi: Orthophosphate; PPDK: Pyruvate,Pi dikinase; PPFD: Photosynthetic photon flux density, μmol quanta m-2 s-1; PPi: Pyrophosphate; PS I: Photosystem I; PS II: Photosystem II; rbcL: Rubisco large subunit mRNA; rbcL: Rubisco large subunit protein; RbcS: Rubisco small subunit mRNA; RbcS: Rubisco small subunit protein; RLSB: rbcL RNA S1-binding domain protein; Rubisco: Ribulose bisphosphate carboxylase-oxygenase; TRX: Thioredoxin.
Competing interests
• In the past five years or in the future, no authors have received or will receive any reimbursements, fees, funding, or salary from an organization that may gain or lose financially from publication of this manuscript. No organization is financing this manuscript.
• No author holds any stocks or shares in an organization that, either now or in the future, may gain or lose financially from the publication of this manuscript.
• No author is currently applying for any patents relating to the content of the manuscript. No author has received reimbursements, fees, funding, or salary from an organization that holds or has applied for patents relating to the content of the manuscript.
• No author has any financial or non-financial competing interests to declare in relation to this manuscript.
Authors’ contributions
JR carried out the assembly of constructs, biolistic and protoplast transformation, participated in the immunolocalization analysis and RLSB sequence analysis, performed the DAB histochemical analysis and drafted the manuscript. PY carried out the immunolocalization analysis, assisted in RLSB sequence analysis and editing of manuscript. JB participated in design and coordination, assisted with the immunolocalization analysis, and helped in writing the manuscript. TO participated in the design of the study. GE conceived the study, participated in the design and coordination, and helped draft the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
List of constructs and summary of results from GFP expression. Table S2. List of primer sequences used for this study.
Biolistic results with puc18 spGFP.
Biolistic results with PPDK-180 spGFP.
Biolistic results with RbcS-273 spGFP.
Biolistic results with RLSB spGFP.
Representative image for quantification of GFP fluorescence from RbcS-FL spGFP and RbcS-FL roGFP2.
DNA sequences used in constructs.
Contributor Information
Josh Rosnow, Email: jrosnow@wsu.edu.
Pradeep Yerramsetty, Email: pradeepk@buffalo.edu.
James O Berry, Email: camjob@buffalo.edu.
Thomas W Okita, Email: okita@wsu.edu.
Gerald E Edwards, Email: edwardsg@wsu.edu.
Acknowledgements
This material is based upon work supported by the National Science Foundation under funds MCB #1146928 to GEE, and USDA/NRI Grant 2008–01070 to J.O.B. Additional thanks to Ray Collier for help in assembling and providing the constructs, Michael Knoblauch, Valerie Lynch-Holm, Christine Davitt in the Franceschi Microscopy Center for discussions and assistance with imaging, Nuria Koteyeva for parafin samples, and Chuck Cody for plant growth management. Travel support for JR to the University of New York – Buffalo was provided by a Betty Higginbotham grant through the School of Biological Sciences at WSU.
References
- von Caemmerer S, Furbank RT. The C4 pathway: an efficient CO2 pump. Photosynth Res. 2003;77:191–207. doi: 10.1023/A:1025830019591. [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F. Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 2012;63:19–47. doi: 10.1146/annurev-arplant-042811-105511. [DOI] [PubMed] [Google Scholar]
- Edwards G, Walker D. C3, C4: Mechanism, and Cellular and Environmental Regulation, of Photosynthesis. London: Blackwell Scientific Publications; 1983. [Google Scholar]
- Edwards GE, Voznesenskaya EV. In: C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Advances in Photosynthesis and Respiration. Volume 32. Raghavendra AS, Sage RF, editor. The Netherlands: Springer Dordrecht; 2011. C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants; pp. 29–61. [Google Scholar]
- Langdale JA. C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell. 2011;23:3879–3892. doi: 10.1105/tpc.111.092098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-L, Long JJ, Hotchkiss T, Berry JO. C4 photosynthetic gene expression in light- and dark-grown amaranth cotyledons. Plant Physiol. 1993;102:1085–1093. doi: 10.1104/pp.102.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q. et al. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell. 2010;22:3509–3542. doi: 10.1105/tpc.110.079764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne BC, Dee AM, Edwards GE. Photosynthesis in mesophyll protoplasts and bundle sheath cells of various type of C4 plants. III. Fluorescence emission spectra, delayed light emission, and P700 Content. Z Pflanzenphysiol. 1974;74:275–291. [Google Scholar]
- Ueno O, Yoshimura Y, Sentoku N. Variation in the activity of some enzymes of photorespiratory metabolism in C4 grasses. Ann Bot. 2005;96:863–869. doi: 10.1093/aob/mci238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, Van Wijk KJ. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 2010;15:1219–1250. doi: 10.1104/pp.109.152694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer MU, Santore UJ, Westhoff P. Differential accumulation of the 10-, 16- and 23-kDa peripheral components of the water-splitting complex of photosystem II in mesophyll and bundle-sheath chloroplasts of the dicotyledonous C4 plant Flaveria trinervia (Spreng.) C. Mohr Planta. 1992;186:304–312. doi: 10.1007/BF00196260. [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Pyankov VI, Edwards GE. Anatomy, chloroplast structure and compartmentation of enzymes relative to photosynthetic mechanisms in leaves and cotyledons of species in the tribe Salsoleae (Chenopodiaceae) J Exp Bot. 1999;50(341):1779–1795. [Google Scholar]
- Hibberd JM, Covshoff S. The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 2010;61:181–207. doi: 10.1146/annurev-arplant-042809-112238. [DOI] [PubMed] [Google Scholar]
- Williams BP, Aubry S, Hibberd JM. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 2012;17:213–220. doi: 10.1016/j.tplants.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Schulze S, Mallmann J, Burscheidt J, Koczor M, Streubel M, Bauwe H, Gowik U, Westhoff P. Evolution of C4 photosynthesis in the genus Flaveria: establishment of a photorespiratory CO2 pump. Plant Cell. 2013;25:2522–2535. doi: 10.1105/tpc.113.114520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Brautigam A, Schluter U, Denton AK, Colmsee C, Scholz U, Fahnenstich H, Pieruschka R, Rascher U, Sonnewald U, Weber APM. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell. 2011;23:4208–4220. doi: 10.1105/tpc.111.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards GE, Franceschi VR, Voznesenskaya EV. Single-Cell C4 photosynthesis versus the dual-cell (Kranz) paradigm. Annu Rev Plant Physiol Plant Mol Biol. 2004;55:173–196. doi: 10.1146/annurev.arplant.55.031903.141725. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ. The C4 plant lineages of planet Earth. J Exp Bot. 2011;62:3155–3169. doi: 10.1093/jxb/err048. [DOI] [PubMed] [Google Scholar]
- Akhani H, Chatrenoor T, Dehghani M, Khoshravesh R, Mahdavi P, Matinzadeh Z. A new species of Bienertia (Chenopodiaceae) from Iranian salt deserts: a third species of the genus and discovery of a fourth terrestrial C4 plant without Kranz anatomy. Plant Biosystems - An International Journal Dealing with all Aspects of Plant Biology: Official Journal of the Societa Botanica Italiana. 2012.
- Freitag H, Stichler W. A remarkable new leaf type with unusual photosynthetic tissue in a central asiatic genus of chenopodiaceae. Plant Biol. 2000;2:154–160. doi: 10.1055/s-2000-9462. [DOI] [Google Scholar]
- Offermann S, Okita TW, Edwards GE. Resolving the compartmentation and function of C4 photosynthesis in the single-cell C4 species Bienertia sinuspersici. Plant Physiol. 2011;155:1612–1628. doi: 10.1104/pp.110.170381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Chuong SDX, Akhani H, Edwards GE, Franceschi VR. Differentiation of cellular and biochemical features of the single-cell C4 syndrome during leaf development in Bienertia cycloptera (Chenopodiaceae) Am J Bot. 2005;91:1784–1795. doi: 10.3732/ajb.92.11.1784. [DOI] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011;155:27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Edwards GE, Kiirats O, Artyusheva EG, Franceschi VR. Development of biochemical specialization and organelle partitioning in the single-cell C4 system in leaves of Borszczowia aralocaspica (Chenopodiaceae) Am J Bot. 2003;90:1669–1680. doi: 10.3732/ajb.90.12.1669. [DOI] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D. Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc Natl Acad Sci. 2007;104:6466–6471. doi: 10.1073/pnas.0610586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SM, Patel M, Yerramsetty P, Mure CM, Zielinski AM, Bruenn JA, Berry JO. A novel RNA binding protein affects rbcL gene expression and is specific to bundle sheath chloroplasts in C4 plants. BMC Plant Biol. 2013;13:138–162. doi: 10.1186/1471-2229-13-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L, Williams-Carrier R, Wostrikoff K, Belcher S, Barkan A, Stern DB. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell. 2012;24:3435–3446. doi: 10.1105/tpc.112.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S-B, Kim K-H, Kavakli IH, Lee S-K, Okita TW. Transcriptional expression characteristics and subcellular localization of ADP-glucose pyrophosphorylase in the oil plant Perilla frutescens. Plant Cell Physiology. 2001;42:146–153. doi: 10.1093/pcp/pce019. [DOI] [PubMed] [Google Scholar]
- Park J, Okita TW, Edwards GE. Expression profiling and proteomic analysis of isolated photosynthetic cells of the non-Kranz C4 species Bienertia sinuspersici. Funct Plant Biol. 2010;37:1–13. doi: 10.1071/FP09074. [DOI] [Google Scholar]
- Seibel NM, Eljouni J, Nalaskowski MM, Hampe W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem. 2007;368:95–99. doi: 10.1016/j.ab.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- Furbank RT, von Caemmerer S, Sheehy J, Edwards G. C4 rice: a challenge for plant phenomics. Funct Plant Biol. 2009;36:845–856. doi: 10.1071/FP09185. [DOI] [PubMed] [Google Scholar]
- Pfundel E, Neubohn B. Assessing photosystem I and II distribution in leaves from C4 plants using confocal laser scanning microscopy. Plant Cell Environ. 1999;22:1569–1577. doi: 10.1046/j.1365-3040.1999.00521.x. [DOI] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Kiirats O, Artyusheva EG, Freitag H, Edwards GE. Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae) Plant J. 2002;31:649–662. doi: 10.1046/j.1365-313X.2002.01385.x. [DOI] [PubMed] [Google Scholar]
- Koteyeva NK, Voznesenskaya EV, Berry JO, Chuong SDX, Franceschi VR, Edwards GE. Development of structural and biochemical characteristics of C4 photosynthesis in two types of Kranz anatomy in genus Suaeda (family Chenopodiaeae) J Exp Bot. 2011;62:3197–3212. doi: 10.1093/jxb/err021. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Shigeru S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011;155:93–100. doi: 10.1104/pp.110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynth Res. 2008;98:551–564. doi: 10.1007/s11120-008-9349-3. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Akagi H, Kamasawa N, Osumi M, Honda H. Aberrant chloroplasts in transgenic rice plants expressing a high level of maize NADP-dependent malic enzyme. Planta. 2000;211:265–274. doi: 10.1007/s004250000282. [DOI] [PubMed] [Google Scholar]
- Offermann S, Okita TW, Edwards GE. How do single cell C4 species form dimorphic chloroplasts? Plant Signal Behav. 2011;6:762–765. doi: 10.4161/psb.6.5.15426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung S-C, Yanagisawa M, Chuong SDX. Protoplast isolation and transient gene expression in the single-cell C4 species, Bienertia sinuspersici. Plant Cell Rep. 2010;30:473–484. doi: 10.1007/s00299-010-0953-2. [DOI] [PubMed] [Google Scholar]
- Chuong SDX, Franceschi VR, Edwards GE. The cytoskeleton maintains organelle partitioning required for single-cell C4 photosynthesis in Chenopodiaceae species. Plant Cell. 2006;18:2207–2223. doi: 10.1105/tpc.105.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Sorenson R, Juntawong P. Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 2009;14:443–454. doi: 10.1016/j.tplants.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Doroshenk KA, Satoh-Cruz M, Nagamine A, Katsube-Tanaka T, Ogawa M, Kumamaru T, Satoh H, Okita TW. RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J. 2012;70:471–479. doi: 10.1111/j.1365-313X.2011.04880.x. [DOI] [PubMed] [Google Scholar]
- Park S-H, Chung PJ, Juntawong P, Bailey-Serres J, Kim YS, Jung H, Bang SW, Kim Y-K, Choi YD, Kim J-K. Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol. 2012;159:1111–1124. doi: 10.1104/pp.112.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Corey AC, Yin L-P, Ali S, Taylor WC, Berry JO. Untranslated regions from C4 Amaranth AhRbcS1 mRNAs confer translational enhancement and preferential bundle sheath cell expression in transgenic C4 Flaveria bidentis. Plant Physiol. 2004;136:3550–3561. doi: 10.1104/pp.104.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell CA, Brown NJ, Liu Z, Pflug A, Gowik U, Westhoff P, Hibberd JM. Agrobacterium tumefaciens - mediated transformation of Cleome gynandra L., a C4 dicotyledon that is closely related to Arabidopsis thaliana. J Exp Bot. 2010;61:1311–1319. doi: 10.1093/jxb/erq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Purcell M, Zucchi P, Helentjaris T, Bogorad L. TRM1, a YY1-like suppressor of rbcS-m3 expression in maize mesophyll cells. Proc Natl Acad Sci. 2001;98:2295–2300. doi: 10.1073/pnas.041610098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung S-C, Chuong SDX. A transit peptide-like sorting signal at the C terminus directs the Bienertia sinuspersici preprotein receptor Toc159 to the chloroplast outer membrane. Plant Cell. 2012;24:1560–1578. doi: 10.1105/tpc.112.096248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Abell BM. Chloroplast envelope protein targeting fidelity is independent of cytosolic components in dual organelle assays. Frontiers Plant Sci. 2012;3:1–11. doi: 10.3389/fpls.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Reddick LE, McWilliams DR, Campbell IM, Bruce BD. Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell. 2012;24:3040–3059. doi: 10.1105/tpc.112.098327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- H-m L, Chiu C-C. Protein transport into chloroplasts. Annu Rev Plant Physiol Plant Mol Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- Stengel A, Benz PJ, Buchanan BB, Soll J, Bolter B. Preprotein import into chloroplasts via the Toc and Tic complexes is regulated by redox signals in Pisum sativum. Mol Plant. 2009;2:1181–1197. doi: 10.1093/mp/ssp043. [DOI] [PubMed] [Google Scholar]
- Stengel A, Benz P, Balsera M, Soll J, Bolter B. TIC62 redox-regulated translocon composition and dynamics. J Biol Chem. 2008;283:6656–6667. doi: 10.1074/jbc.M706719200. [DOI] [PubMed] [Google Scholar]
- Barnes D, Cohen A, Bruick RK, Kantardjieff K, Fowler S, Efuet E, Mayfield SP. Identification and characterization of a novel RNA binding protein that associates with the 5′-untranslated region of the chloroplast psbA mRNA. Biochemistry. 2004;43:8541–8550. doi: 10.1021/bi035909j. [DOI] [PubMed] [Google Scholar]
- McCormac DJ, Litz H, Wang J, Gollnick PD, Berry JO. Light-associated and processing-dependent protein binding to 5′ regions of rbcL mRNA in the chloroplast of a C4 plant. J Biol Chem. 2001;276:3476–3483. doi: 10.1074/jbc.M009236200. [DOI] [PubMed] [Google Scholar]
- Zerges W, Rochaiz J-D. Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 1998;140:101–110. doi: 10.1083/jcb.140.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JO, Yerramsetty P, Zielinski AM, Mure CM. Photosynthetic gene expression in higher plants. Photosynth Res. 2013;117:91–120. doi: 10.1007/s11120-013-9880-8. [DOI] [PubMed] [Google Scholar]
- Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Irihimovitch V, Knopf JA, Cohen I, Orr-Dahan I, Nahum E, Keasar C, Shapira M. RNA binding activity of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Chlamydomonas reinhardtii. J Biol Chem. 2004;279:10148–10156. doi: 10.1074/jbc.M308602200. [DOI] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finzaai G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Woollman F-A, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidospsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP, Sawers RJH, Mant A, Langdale JA. BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell. 1999;11:849–864. doi: 10.1105/tpc.11.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Trivedi PK, Sethuraman A, Mehrotra R. The rbcL gene of Populus deltoides has multiple transcripts and is redox-regulated in vitro. J Plant Physiol. 2011;168:466–473. doi: 10.1016/j.jplph.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Wostrikoff K, Clark A, Sato S, Clemente T, Stern D. Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type-specific accumulation. Plant Physiol. 2012;160:419–432. doi: 10.1104/pp.112.195677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J, Garcia-Murria MJ, Marin-Navarro J. Redox modulation of rubisco conformation and activity through its cysteine residues. J Exp Bot. 2008;59:1605–1614. doi: 10.1093/jxb/erm310. [DOI] [PubMed] [Google Scholar]
- Cohen I, Sapir Y, Shapira M. A conserved mechanism controls translation of Rubisco large subunit in different photosynthetic organisms. Plant Physiol. 2006;141:1089–1097. doi: 10.1104/pp.106.079046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Lers A, Heifetz PB, Irihimovitch V, Osmond CB, Gillham NW, Boynton JE. Differential regulation of chloroplast gene expression in Chlamydomonas reinhardtii during photoacclimation: light stress transiently suppresses synthesis of the Rubisco LSU protein while enhancing synthesis of the PS II D1 protein. Plant Mol Biol. 1997;33:1001–1011. doi: 10.1023/A:1005814800641. [DOI] [PubMed] [Google Scholar]
- Irihimovitch V, Shapira M. Glutathione redox potential modulated by reactive oxygen species regulates translation of rubisco large subunit in the chloroplast. J Biol Chem. 2000;275:16289–16295. doi: 10.1074/jbc.275.21.16289. [DOI] [PubMed] [Google Scholar]
- Li S, Lu W, Li G-F, Gong Y-D, Zhao N-M, Zhang R-X, Zhou H-M. Interaction of hydrogen peroxide with ribulose-1,5-bisphosphate carboxylase/oxygenase from rice. Biochem Mosc. 2004;69:1136–1142. doi: 10.1023/b:biry.0000046888.45252.67. [DOI] [PubMed] [Google Scholar]
- Patel M, Berry JO. Rubisco gene expression in C4 plants. J Exp Bot. 2008;59:1625–1634. doi: 10.1093/jxb/erm368. [DOI] [PubMed] [Google Scholar]
- Berry JO, Patel M, Zielinski A. In: C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Advances in Photosynthesis and Respiration. Volume 32. Raghavendra AS, Sage RF, editor. The Netherlands: Springer Dordrecht; 2011. C4 gene expression in mesophyll and bundle sheath cells; pp. 221–250. [Google Scholar]
- Chiu W-l, Niwa Y, Zeng W, Hirano T, Kobayashi H, Shenn J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Mathur J, Koncz C. PEG-Mediated Protoplast Transformation with Naked DNA. Totowa: Humana Press Inc.; 2005. [DOI] [PubMed] [Google Scholar]
- Berry JO, Nikolau BJ, Carr JP, Klessig DF. Transcriptional and post-transcriptional regulation of ribulose 1,5-bisphosphate carboxylase gene expression in light- and dark-grown amaranth cotyledons. Mol Cell Biol. 1985;5:2238–2246. doi: 10.1128/mcb.5.9.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnow J, Offermann S, Park J, Okita TW, Tarlyn N, Dhingra A, Edwards GE. In vitro cultures and regeneration of Bienertia sinuspersici (Chenopodiaceae) under increasing concentrations of sodium chloride and carbon dioxide. Plant Cell Rep. 2011;30:1541–1553. doi: 10.1007/s00299-011-1067-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of constructs and summary of results from GFP expression. Table S2. List of primer sequences used for this study.
Biolistic results with puc18 spGFP.
Biolistic results with PPDK-180 spGFP.
Biolistic results with RbcS-273 spGFP.
Biolistic results with RLSB spGFP.
Representative image for quantification of GFP fluorescence from RbcS-FL spGFP and RbcS-FL roGFP2.
DNA sequences used in constructs.