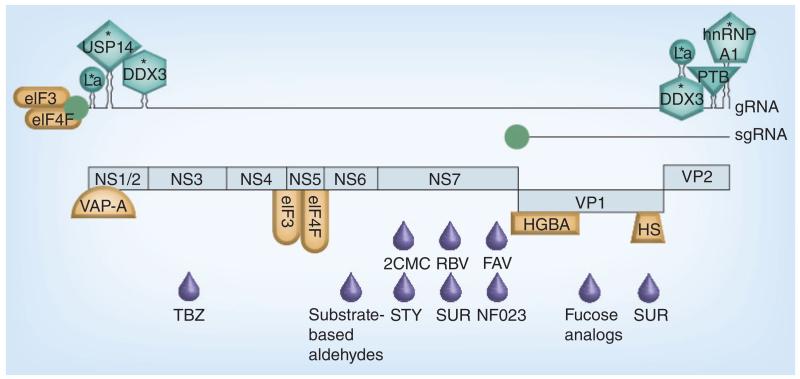
Figure 1. Overview of the known and possible antiviral targets for noroviruses.
The norovirus gRNA and sgRNA are shown as a larger and a shorter line, respectively, each linked to a VPg molecule (green circle). RNA secondary structures known to form at the extremities of the genome are schematically drawn. The different open reading frames expressed are represented below the genome as light blue boxes. Open reading frame 1 is subdivided into different segments corresponding to the different nonstructural proteins released after proteolysis: NS1/2, NTPase (NS3), NS4, VPg (NS5), protease (NS6pro) and RNA-dependent RNA polymerase (NS7pol). Host factors identified to bind to the viral genome extremities are represented as different green shapes. Host factors interacting with viral proteins are represented as different orange shapes. Asterisks highlight host factors for which small molecule inhibitors are available, and their inhibition by these drugs, or downregulation by RNAi, result in decreased norovirus replication. Note that the components shown to interact with the viral RNA genome may interact indirectly via an intermediate protein or proteins. Antiviral compounds targeting different viral proteins are represented below each corresponding region of the genome as purple drop-shaped figures.
2CMC: 2′-C-methylcytidine; FAV: Favipiravir; gRNA: Genomic RNA; RBV: Ribavirin; sgRNA: Subgenomic RNA; STY: Styrylchromones; SUR: Suramin; TBZ: Thiazolobenzimidazoles.