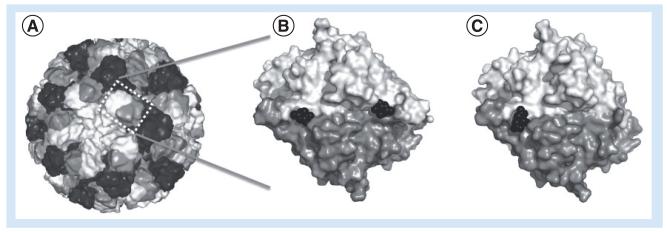
Figure 3. Structure of the viral capsid in complex with natural fucose substrate and citrate inhibitor.
(A) Structure of human norovirus capsid: 60 asymmetric units are assembled to form a mature capsid. Each asymmetric unit contains three copies of VP1 represented in a different grayscale. (B) Top view of a human norovirus VP1 dimer bound to two fucose residues (black), the natural binding substrates of the capsid. (C) A similar representation as in (B), but illustrating the interaction with citrate, a fucose analog inhibitor, interacting with the fucose-binding pocket. For the modelling of the full norovirus capsid, the program Protein Workshop and the structure available for Norwalk virus (PDB: 1IHM) has been used. PyMol program has been used for the representation of VP1 dimers bound to fucose and citrate (human norovirus Vietnam strain 026, PDBs: 3ONY and 3RY8).