Abstract
Plant growth and productivity are greatly affected by drought, which is likely to become more threatening with the predicted global temperature increase. Understanding the genetic architecture of complex quantitative traits and their interaction with water availability may lead to improved crop adaptation to a wide range of environments. Here, the genetic basis of 20 physiological and morphological traits is explored by describing plant performance and growth in a Brassica rapa recombinant inbred line (RIL) population grown on a sandy substrate supplemented with nutrient solution, under control and drought conditions. Altogether, 54 quantitative trait loci (QTL) were identified, of which many colocated in 11 QTL clusters. Seventeen QTL showed significant QTL–environment interaction (Q×E), indicating genetic variation for phenotypic plasticity. Of the measured traits, only hypocotyl length did not show significant genotype–environment interaction (G×E) in both environments in all experiments. Correlation analysis showed that, in the control environment, stomatal conductance was positively correlated with total leaf dry weight (DW) and aboveground DW, whereas in the drought environment, stomatal conductance showed a significant negative correlation with total leaf DW and aboveground DW. This correlation was explained by antagonistic fitness effects in the drought environment, controlled by a QTL cluster on chromosome A7. These results demonstrate that Q×E is an important component of the genetic variance and can play a great role in improving drought tolerance in future breeding programmes.
Key words: Antagonistic fitness effect, Brassica rapa, drought, genotype–environment interaction, plasticity, root/shoot ratio, stomatal conductance.
Introduction
Plant growth is greatly affected by environmental abiotic stresses, of which drought is the most common factor impeding crop productivity. Drought is likely to become more threatening with the predicted global temperature increase (Smith and De Smet, 2012). Three categories of plant adaptive strategies to drought have been recognized: drought escape by early flowering, drought tolerance via increasing water use efficiency and drought avoidance via reduced transpiration and increasing water uptake (Levitt, 1972).
Evaluating those responses in many genotypes in several environments may show phenotypic plasticity, which is defined as the ability of an individual organism to alter its physiology/morphology in response to changes in environmental conditions (Schlichting, 1986). When this plasticity differs between genotypes (i.e. when there is genetic variation for it), it is classified as genotype–environment interaction (G×E) (Via and Lande, 1985). Understanding G×E better will provide a solid foundation for genetic improvement of stable crop productivity and will help to identify superior and stable alleles/genotypes across different environments (Zhang et al., 2010). The genetic basis of the observed G×E can be identified by genetically dissecting plant physiological and morphological responses to environments via quantitative trait loci (QTL). This specifies the genetic component of G×E and is expressed as QTL–environment interaction (Q×E) (Malosetti et al., 2004; Boer et al., 2007; Tardieu, 2013). Different QTL effects can occur if the allele underlying the QTL is strongly expressed in one environment but weakly in another, or if the allele has opposite effects on the same trait in different environments (Mackay, 2001; Sukhwinder et al., 2012). A QTL for which one allele has opposite (pleiotropic) effects on the phenotype in two different environments can lead to fitness trade offs, elevating fitness in one environment but depressing it in the other environment. Trade offs can be maintained in nature (e.g. by antagonistic pleiotropy), when alleles at a locus underlying a fitness component show clear home-site advantages (Rose, 1982; Anderson et al., 2013). Therefore, considering such antagonistic fitness effects is crucial while selecting for desirable QTL during marker-assisted breeding programmes.
To facilitate improving marker-assisted breeding programmes, a model crop plant is required. The Brassica genus has the smallest genome size, the complete genome sequence of Brassica rapa (Wang et al., 2011), close relationships with the plant model species Arabidopsis thaliana and genome analysis tools, provided in the Brassica database (BRAD) (Cheng et al., 2011), so B. rapa is a useful dicot model crop for genetic and comparative studies.
The present study focused on drought avoidance, which enables plants to maintain a high fitness level in drought conditions. Therefore, Q×E on growth-related traits were investigated in a B. rapa recombinant inbred line (RIL) population grown on a sandy substrate under control and drought environments. This work identified several QTL for main effects and Q×E and found an antagonistic fitness effect for a stomatal conductance/shoot biomass QTL, with the same allele reducing stomatal conductance under drought and increasing it under normal watering conditions, while contributing to higher shoot biomass in both environments.
Materials and methods
Plant material and experimental setup
The RIL population (F7) used here was previously developed by this study group from a cross between a Yellow Sarson (R-o-18) (♂) and a Caixin type (L58) (♀) and genotyped with 270 markers (Bagheri et al., 2012). The RIL population was screened three times under control (continuous watering for 3 weeks) and drought (normal watering for 1 week, then plants were left to dry out) environments. In all screens, plants were grown in 13-cm-deep square black plastic pots. Each pot was filled with 1.5kg dried river sand and all pots were watered until saturation with 1100ml nutrient solution (1, 1.1, 5.9 mmol l–1, N, P, and K respectively). The same nutrient solution was used for watering plants every 2 days. Two seeds were sown per pot and 4 days after germination, seedlings were thinned to one per pot. Seven days after germination, watering was withheld as drought treatment, while the control treatment was continuously watered.
Initially, a pilot experiment was performed using 30 randomly selected RILs and both parental lines, with three replications per genotype per environment, to test if the drought treatment would reveal significant differences between RILs and between the two environments regarding total leaf fresh and dry weight. Subsequently, a full RIL screening experiment was performed in which 140 RILs and both parents were phenotyped for the 20 studied traits under both environments with three replications per RIL and six replications per parental line per treatment. Finally a QTL reproducibility experiment was performed to confirm the different phenotypes for contrasting alleles at four identified QTL by screening 27 RILs selected for their discriminating genotypes, with three replicates per RIL per environment.
All experiments were carried out under controlled greenhouse conditions under a16/8 light/dark cycle (22.3/20.3°C, mean relative humidity 77.8/81.3%). The experimental setup involved a complete randomized block design with one plant per RIL and two replicates for each parent per block.
Plant phenotyping
In the full RIL screening and QTL reproducibility experiments, 20 traits were analysed under control and drought environments. These traits were chosen as the ones describing as best as possible the different aspects of plant performance. Directly before harvesting, when less than 5% of plants had visible flower primordia, the number of leaves was counted. Chlorophyll content was measured (only in the full RIL screening experiment) using a SPAD-502 chlorophyll meter (Minolta, Japan). For this measurement the average of three leaves per plant per replication per treatment was taken. Leaf stomatal conductance was measured using a leaf porometer (Decagon Devices, USA) for one fully expanded leaf per plant per replication (either the 3rd or 4th leaf). Thereafter, total leaf fresh weight (LFW) and dry weight (LDW) and the dry weight of the 3rd and 4th (i.e. fully expanded) leaves (3,4DW) was measured. Dry weights were determined after drying plant materials at 65°C for 4–5 days until weight constancy.
Leaf area (LA) of the 3rd and 4th leaves was measured using a Licor LI-3100 (Licor, Lincoln, NE, USA), and subsequently their combined specific leaf area (SLA) was calculated as LA divided by 3,4DW, as well as the dry weight ratio between 3,4DW and LDW. Hypocotyl length was measured using a ruler, and hypocotyl DW (HDW) was determined. The shoot DW (SDW) was calculated as the sum of LDW and HDW.
Subsequently, root systems were washed carefully to remove adhering sand, placed in a plastic tray filled with water, spread and scanned with a flatbed scanner. From this, the total root system length (RL), root volume (RV), and root diameter (RD) were measured using WinRhizo (Regent Instruments, Quebec, Canada). This was used to calculate the RL-to-SDW ratio (RL/SDW), which illustrates the aboveground matter that is supported by a given RL.
Thereafter, roots were dried to measure root DW (RDW) and to calculate the root-to-shoot DW ratio (R/S). Similarly, to indicate the relative investment in shoots or roots, the shoot-to-total plant (shoot + root) DW ratio was calculated (S/SR), for which total plant DW was calculated as the sum of SDW and RDW. Finally, the leaf water content (LWC) was calculated as (LFW – LDW) / LDW.
Statistical and quantitative trait loci analysis
Statistical analysis was performed on raw data of each experiment using GenStat for Windows 15th edition (VSN International, Hemel Hempstead, UK). Analysis of variance (ANOVA) was used to test the significance difference between treatments, lines, and interaction (G×E). Heritability was estimated as implemented in GenStat. In the linear mixed model, genotypes were fitted as random and blocks as fixed. The generalized heritability measure used, as described by Cullis et al. (2006), and in a more general context by Welham et al. (2010), is given by:
where the set of predicted genotype means (Best Linear Unbiased Predictors) are g 1 … g N with prediction error variance pev(g i) and estimated genetics variance component . Pearson correlations were calculated using GenStat.
Data from the 20 traits analysed in the full RIL screening experiment were used for QTL mapping using a multienvironment analysis (MEA) approach, which accounts for G×E, as implemented in the QTL library in GenStat. A step size of 10 cM, a minimum cofactor proximity of 50 cM, a minimum separation of selected QTL of 30 cM, and a threshold of –log10P = 2.8 were used for QTL analysis. Following the mixed-model approach described by (Malosetti et al., 2004; Boer et al., 2007), first the whole genome was scanned using simple interval mapping and then, based on that, cofactors were selected for two rounds of composite interval mapping. Thereafter, a final QTL model was selected using backward selection on the selected cofactors, where it estimated the allelic effect of each of QTL in each environment, the effect of Q×E, and the explained phenotypic variance of each QTL per environment. In addition to determining phenotypic plasticity as Q×E, a second method to determine plasticity QTL was used as described by (Tétard-Jones et al., 2011), by QTL mapping the difference in the mean phenotypic values per line between treatments.
Confirming reproducibility of four QTL clusters
To confirm the reproducibility of the major QTL detected in the full RIL screening experiment, this work selected four QTL clusters: on chromosome 3 at 38–42 cM, on chromosome 7 at 30–40 cM, on chromosome 8 at 85–95 cM, and on chromosome 9 at 70–84 cM. The whole population was genetically classified into 16 groups based on all possible allelic combinations at the four selected QTL. Thereafter, for every tested QTL, phenotypic data of RILs with contrasting genotypes for one QTL, but similar genotypes for the other QTL, were compared. For example, to test for the QTL on chromosome 3, ANNN RILs were compared with BNNN RILs in paired groups, so AAAA with BAAA, ABAA with BBAA, ABBA with BBBA, etc. The 27 RILs with the highest and lowest average values at each tested QTL were selected and grown as described. For all measured traits, a correlation analysis between traits measured in the control environments and traits measured in the drought environments of the full RIL screening experiment and the QTL reproducibility experiments was used to test for a significantly similar response to the treatment as a confirmation of the level of reproducibility.
Results
Phenotyping the RIL population
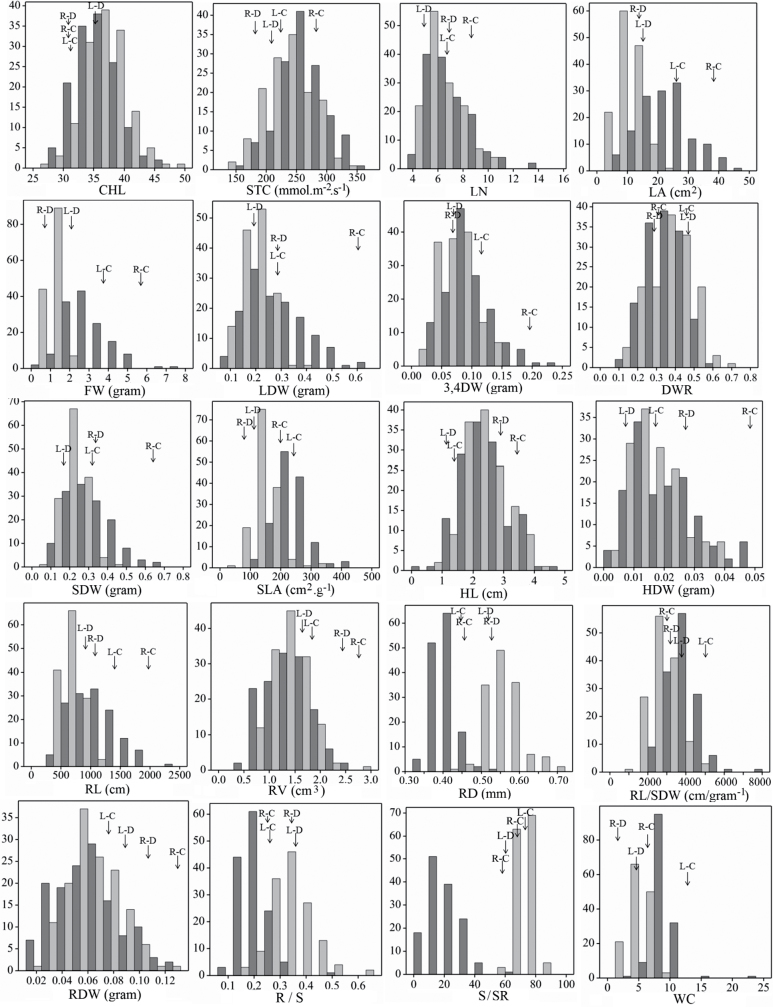
The results obtained from the pilot experiment (data not shown) indicated there was ample phenotypic variation for drought response, which justified phenotyping the whole RIL population. A total of 20 traits related to growth and performance of plants were analysed under control and drought environments. Fig. 1 shows the frequency distributions of the measured traits over the whole population. Transgression beyond both parental lines was observed for most of the traits except for root volume, RDW, HDW, SDW, S/SR, and LWC, where transgression was only in one direction.
Fig. 1.
Frequency distributions of the non-normalized trait values for the L58 × R-o-18 recombinant inbred line population under control (C) (dark grey), and drought (D) (light grey) conditions. Vertical axes indicate the number of lines per trait value class, and horizontal axes indicate the different trait value classes. L, L58; R, R-o-18. Trait abbreviations are given with Table 1.
The drought treatment decreased fresh weight, leaf number, leaf area, LDW, root length, and stomatal conductance and increased R/S (Fig. 1, Table 1). For stomatal conductance, the reduction in the L58 parent was minor and not significant, as was also the case for some of the RILs.
Table 1.
Parental line means for the analysed traits and performance of the phenotyped RIL population under control and drought conditionsFor all traits, three replicates were measured. Min and Max indicate the lowest and highest RIL values; mean and standard deviation (SD) is for all RILs; h2 indicates broad sense heritability. S, significant; NS, nonsignificant; –, not applicable (ANOVA and heritability cannot be calculated because averages and not replications were used). CHL, chlorophyll content; HDW, hypocotyl dry weight; HL, hypocotyl length; LA, leaf area; LDW, leaf dry weight; LFW, leaf fresh weight; LN, leaf number; LWC, leaf water content; RD, root diameter; RDW, root dry weight; RIL, recombinant inbred line; RL/SDW, root length-to-shoot dry weight ratio; RL, root system length; RV, root volume; R/S, root-to-shoot dry weight ratio; SDW, shoot dry weight; S/SR, shoot-to-plant dry weight ratio; SLA, specific leaf area; STC, stomatal conductance; DWR, 3,4DW vs. LDW ratio; 3,4DW, 3rd and 4th leaf dry weight.
| Trait and treatment | Unit | Parental lines | RIL population | ANOVA 2nd experiment | h2 | ANOVA 3rd experiment | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L58 | R-o-18 | Min | Max | Mean | SDb | Treatment | RILs | G×E | Treatment | RILs | G×E | |||
| CHL with C | 31.84 | 31.45 | 28.20 | 47.45 | 35.77 | 3.47 | S | S | NS | 0.59 | ||||
| CHL with D | 35.14 | 30.96 | 27.40 | 47.70 | 36.87 | 3.58 | 0.50 | |||||||
| STC with C | mmol m–2 s–1 | 237.87 | 266.12 | 173.95 | 364.25 | 265.50 | 37.59 | S | S | S | 0.31 | S | NS | NS |
| STC with D | 220.50 | 165.67 | 125.90 | 340.70 | 231.80 | 39.17 | 0.81 | |||||||
| LN with C | n | 6.67 | 7.50 | 4.50 | 14.00 | 7.24 | 1.75 | S | S | S | 0.56 | S | S | NS |
| LN with D | 5.60 | 6.60 | 3.67 | 10.33 | 6.20 | 1.42 | 0.78 | |||||||
| LA with C | cm2 | 26.38 | 35.79 | 6.33 | 47.91 | 23.73 | 8.47 | S | S | NS | 0.26 | S | S | S |
| LA with D | 11.87 | 10.04 | 1.28 | 22.24 | 9.23 | 3.83 | 0.46 | |||||||
| LFW with C | g | 3.68 | 5.11 | 0.772 | 7.458 | 2.994 | 1.136 | S | NS | NS | 0.44 | S | S | S |
| LFW with D | 1.21 | 0.77 | 0.268 | 2.046 | 0.979 | 0.324 | 0.36 | |||||||
| LDW with C | g | 0.276 | 0.619 | 0.087 | 0.637 | 0.293 | 0.114 | S | S | NS | 0.31 | S | S | S |
| LDW with D | 0.196 | 0.275 | 0.060 | 0.370 | 0.193 | 0.057 | 0.37 | |||||||
| 3,4DW with C | g | 0.112 | 0.194 | 0.032 | 0.231 | 0.099 | 0.039 | S | S | NS | 0.48 | S | S | S |
| 3,4DW with D | 0.082 | 0.081 | 0.017 | 0.150 | 0.069 | 0.029 | 0.57 | |||||||
| DWR with C | 0.406 | 0.314 | 0.106 | 0.561 | 0.356 | 0.096 | – | – | – | – | – | – | – | |
| DWR with D | 0.419 | 0.295 | 0.098 | 0.670 | 0.366 | 0.117 | – | |||||||
| SDW with C | g | 0.288 | 0.671 | 0.092 | 0.665 | 0.315 | 0.122 | – | – | – | – | – | – | – |
| SDW with D | 0.205 | 0.300 | 0.074 | 0.408 | 0.210 | 0.062 | – | |||||||
| SLA with C | cm2 g–1 | 235.03 | 184.48 | 139.00 | 426.35 | 246.20 | 52.43 | – | – | – | – | – | – | – |
| SLA with D | 144.45 | 123.95 | 35.42 | 325.75 | 137.60 | 42.89 | – | |||||||
| HL with C | cm | 1.60 | 3.40 | 0.500 | 4.550 | 2.439 | 0.738 | S | S | NS | 0.90 | S | S | NS |
| HL with D | 1.10 | 2.78 | 0.733 | 4.300 | 2.373 | 0.692 | 0.91 | |||||||
| HDW with C | g | 0.012 | 0.052 | 0.003 | 0.048 | 0.021 | 0.011 | S | S | NS | 0.33 | S | S | S |
| HDW with D | 0.009 | 0.025 | 0.004 | 0.038 | 0.017 | 0.008 | 0.60 | |||||||
| RL with C | cm | 1398.50 | 1970.45 | 304.40 | 2300.00 | 1078.00 | 388.60 | S | S | S | 0.48 | S | S | S |
| RL with D | 741.80 | 940.92 | 254.79 | 1249.64 | 616.60 | 183.40 | 0.48 | |||||||
| RV with C | cm3 | 1.71 | 2.98 | 0.442 | 2.696 | 1.381 | 0.455 | NS | S | NS | 0.38 | NS | S | S |
| RV with D | 1.52 | 2.21 | 0.697 | 2.990 | 1.379 | 0.361 | 0.46 | |||||||
| RD with C | mm | 0.395 | 0.424 | 0.340 | 0.560 | 0.411 | 0.030 | S | S | S | 0.36 | NS | S | S |
| RD with D | 0.500 | 0.550 | 0.429 | 0.686 | 0.548 | 0.046 | 0.26 | |||||||
| RL/SDW with C | cm g–1 | 4849.20 | 2936.90 | 2088.00 | 7298.00 | 3545.00 | 797.40 | – | – | – | – | – | – | – |
| RL/SDW with D | 3616.80 | 3138.90 | 1502.00 | 5525.00 | 3049.00 | 750.30 | – | |||||||
| RDW with C | g | 0.070 | 0.157 | 0.013 | 0.122 | 0.060 | 0.025 | S | S | NS | 0.29 | S | S | S |
| RDW with D | 0.080 | 0.100 | 0.020 | 0.124 | 0.063 | 0.020 | 0.47 | |||||||
| R/S with C | 0.242 | 0.233 | 0.097 | 0.410 | 0.192 | 0.047 | – | – | – | – | – | – | ||
| R/S with D | 0.369 | 0.339 | 0.151 | 0.557 | 0.308 | 0.071 | – | |||||||
| S/SR with C | 77.13 | 74.75 | 2.04 | 60.30 | 21.53 | 10.84 | – | – | – | – | – | – | – | |
| S/SR with D | 69.94 | 68.45 | 55.32 | 93.97 | 70.62 | 4.66 | – | |||||||
| LWC with C | 12.31 | 7.26 | 0.684 | 7.362 | 2.701 | 1.043 | – | – | – | – | – | – | – | |
| LWC with D | 5.16 | 1.80 | 0.110 | 1.765 | 0.786 | 0.300 | – | |||||||
Correlation analysis of all measured traits in this experiment was performed to unveil the genetic and physiological relationships of the various traits (Table 2). The correlations may exist because of similar physiological mechanisms or pleiotropy; however, correlations can also be caused by genetic linkage of loci affecting different traits, which are not physiologically related or pleiotropic. For instance, the analysis showed that, in the control environment, chlorophyll content was positively correlated with root diameter, which is hard to envision being because of pleiotropy.
Table 2.
Pearson correlations for the analysed traits of the L58 × R-o-18 RIL population under control (A) and drought (B) conditionsHighlighted results refer to significant correlations: dark grey, P < 0.01; light grey, P < 0.05. Trait abbreviations are given with Table 1.
| A | |||||||||||||||||||||
| CHL | 1 | - | |||||||||||||||||||
| STC | 2 | -0.017 | - | ||||||||||||||||||
| LN | 3 | -0.075 | 0.115 | - | |||||||||||||||||
| LA | 4 | -0.027 | 0.114 | -0.235 | - | ||||||||||||||||
| LFW | 5 | -0.010 | 0.188 | 0.448 | 0.571 | - | |||||||||||||||
| LDW | 6 | 0.079 | 0.207 | 0.441 | 0.538 | 0.939 | - | ||||||||||||||
| 3,4DW | 7 | 0.096 | 0.129 | -0.125 | 0.851 | 0.668 | 0.737 | - | |||||||||||||
| DWR | 8 | 0.009 | -0.172 | -0.776 | 0.366 | -0.410 | -0.412 | 0.265 | - | ||||||||||||
| SDW | 9 | 0.072 | 0.207 | 0.443 | 0.539 | 0.943 | 0.998 | 0.735 | -0.413 | - | |||||||||||
| SLA | 10 | -0.260 | -0.025 | -0.161 | 0.168 | -0.224 | -0.404 | -0.333 | 0.130 | -0.399 | - | ||||||||||
| HL | 11 | -0.128 | 0.116 | -0.092 | 0.116 | 0.150 | 0.102 | 0.135 | 0.015 | 0.147 | -0.039 | - | |||||||||
| HDW | 12 | -0.023 | 0.139 | 0.329 | 0.387 | 0.696 | 0.671 | 0.493 | -0.296 | 0.719 | -0.228 | 0.562 | - | ||||||||
| RL | 13 | 0.037 | 0.087 | 0.471 | 0.457 | 0.800 | 0.831 | 0.629 | -0.339 | 0.830 | -0.352 | -0.028 | 0.569 | - | |||||||
| RV | 14 | 0.100 | 0.106 | 0.460 | 0.464 | 0.770 | 0.797 | 0.619 | -0.311 | 0.797 | -0.338 | -0.021 | 0.558 | 0.929 | - | ||||||
| RD | 15 | 0.161 | -0.037 | -0.169 | -0.181 | -0.349 | -0.355 | -0.250 | 0.160 | -0.357 | 0.134 | -0.033 | -0.272 | -0.475 | -0.190 | - | |||||
| RL/SDW | 16 | -0.042 | -0.215 | 0.018 | -0.152 | -0.259 | -0.316 | -0.205 | 0.193 | -0.321 | 0.141 | -0.312 | -0.276 | 0.218 | 0.165 | -0.175 | - | ||||
| RDW | 17 | 0.087 | 0.105 | 0.459 | 0.445 | 0.778 | 0.810 | 0.627 | -0.321 | 0.818 | -0.380 | 0.021 | 0.649 | 0.891 | 0.929 | -0.245 | 0.057 | - | |||
| R/S | 18 | 0.068 | -0.086 | 0.140 | -0.061 | -0.053 | -0.102 | -0.035 | 0.082 | -0.084 | -0.008 | -0.088 | 0.123 | 0.264 | 0.358 | 0.096 | 0.660 | 0.441 | - | ||
| S/SR | 19 | 0.084 | 0.161 | 0.410 | 0.483 | 0.830 | 0.905 | 0.684 | -0.370 | 0.904 | -0.393 | 0.041 | 0.623 | 0.820 | 0.790 | -0.324 | -0.190 | 0.819 | 0.057 | - | |
| WC | 20 | -0.224 | -0.112 | -0.078 | -0.044 | -0.039 | -0.336 | -0.289 | 0.114 | -0.320 | 0.559 | 0.106 | -0.066 | -0.227 | -0.228 | 0.086 | 0.299 | -0.238 | 0.247 | -0.327 | - |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
| B | |||||||||||||||||||||
| CHL | 1 | - | |||||||||||||||||||
| STC | 2 | 0.029 | - | ||||||||||||||||||
| LN | 3 | 0.000 | -0.075 | - | |||||||||||||||||
| LA | 4 | -0.035 | -0.107 | -0.320 | - | ||||||||||||||||
| LFW | 5 | 0.013 | -0.080 | 0.131 | 0.571 | - | |||||||||||||||
| LDW | 6 | -0.047 | -0.189 | 0.404 | 0.473 | 0.494 | - | ||||||||||||||
| 3,4DW | 7 | -0.098 | -0.213 | -0.226 | 0.773 | 0.455 | 0.637 | - | |||||||||||||
| DWR | 8 | -0.051 | -0.062 | -0.685 | 0.557 | 0.121 | -0.141 | 0.653 | - | ||||||||||||
| SDW | 9 | -0.066 | -0.187 | 0.405 | 0.463 | 0.473 | 0.995 | 0.618 | -0.160 | - | |||||||||||
| SLA | 10 | 0.072 | 0.141 | -0.122 | 0.305 | 0.123 | -0.119 | -0.283 | -0.218 | -0.103 | - | ||||||||||
| HL | 11 | -0.141 | 0.156 | -0.090 | -0.027 | 0.041 | 0.022 | -0.013 | -0.041 | 0.083 | -0.007 | - | |||||||||
| HDW | 12 | -0.174 | -0.107 | 0.278 | 0.248 | 0.175 | 0.635 | 0.282 | -0.236 | 0.711 | 0.040 | 0.481 | - | ||||||||
| RL | 13 | 0.005 | -0.169 | 0.198 | 0.404 | 0.130 | 0.646 | 0.429 | -0.053 | 0.648 | 0.032 | -0.156 | 0.453 | - | |||||||
| RV | 14 | -0.010 | -0.280 | 0.288 | 0.392 | 0.273 | 0.664 | 0.460 | -0.034 | 0.662 | -0.037 | -0.099 | 0.429 | 0.828 | - | ||||||
| RD | 15 | -0.042 | -0.093 | 0.063 | -0.261 | 0.061 | -0.204 | -0.180 | -0.061 | -0.214 | -0.146 | 0.116 | -0.219 | -0.556 | -0.084 | - | |||||
| RL/SDW | 16 | 0.067 | 0.045 | -0.289 | -0.103 | -0.429 | -0.452 | -0.226 | 0.175 | -0.452 | 0.147 | -0.262 | -0.303 | 0.340 | 0.116 | -0.434 | - | ||||
| RDW | 17 | -0.047 | -0.181 | 0.325 | 0.370 | 0.224 | 0.708 | 0.423 | -0.116 | 0.725 | -0.017 | -0.029 | 0.607 | 0.813 | 0.845 | -0.240 | 0.029 | - | |||
| R/S | 18 | -0.025 | 0.001 | -0.110 | -0.133 | -0.326 | -0.317 | -0.211 | 0.063 | -0.286 | 0.060 | -0.037 | 0.019 | 0.217 | 0.235 | -0.057 | 0.622 | 0.404 | - | ||
| S/SR | 19 | 0.080 | 0.010 | 0.102 | 0.122 | 0.328 | 0.256 | 0.212 | -0.002 | 0.196 | -0.088 | -0.190 | -0.280 | -0.227 | -0.227 | 0.094 | -0.530 | -0.420 | -0.940 | - | |
| WC | 20 | 0.043 | 0.126 | -0.314 | 0.085 | 0.449 | -0.505 | -0.158 | 0.302 | -0.517 | 0.240 | 0.047 | -0.432 | -0.521 | -0.389 | 0.279 | 0.055 | -0.493 | 0.010 | 0.043 | - |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
The correlation observed between root length and S/SR was positive in the control environment (longer roots contributing to relatively more shoots) but negative in the drought environment, indicating a proportionally higher investment in roots. Under both environments, LWC was negatively correlated with SDW, root length, root volume and RDW, while it was positively correlated with LFW and negatively correlated with stomatal conductance under drought conditions. Stomatal conductance was negatively correlated with LDW in the drought environment, but positively correlated under control conditions. In general, plants with longer root systems had higher plant DW.
As expected, all traits measured in control and drought environments showed a positive correlation, except for LWC.
Mapping QTL with main effects and Q×E
In total 54 QTL were mapped for the traits analysed under control and drought environments (Table 3, Fig. 2). Six QTL—STC1, LA4, SLA1, RD3, RL/SDW3, and S/SR2—had opposite allelic effects when comparing both environments. The phenotypic effects of three QTL—LA1, SLA1, and S/SR2—were 9-, 101-, and 15-times higher, respectively, in one environment than the other (Table 3). SLA1 colocated with 3,4DW1, with the alleles increasing the trait values in the control environment from L58 and R-o-18, respectively. Four QTL were mapped for chlorophyll content, of which CHL1, CHL2, and CHL3 showed the highest effect from the L58 allele, while for CHL4 the R-o-18 allele had the highest effect in both environments. Hypocotyl length was mapped to four loci, with the R-o-18 alleles contributing most to increased hypocotyl length. In total, 11 QTL clusters were observed, of which seven comprised at least three colocating QTL (Table 3, Fig. 2).
Table 3.
QTL detected in the L58 × R-o-18 RIL population for the traits described in Table 1, using the multienvironment analysis approachPer trait, QTL are numbered according to chromosome. R 2 is the percentage of total phenotypic variance explained by each QTL. Effects with positive values represent a positive contribution of the R-o-18 allele to the trait value and those with negative values represent a positive contribution of the L58 allele to the trait value. Highlighted results show significant Q×E effects. Ratio refers to the ratios between the effects of each QTL in both environments. Trait abbreviations are given with Table 1.
| Trait | QTL | Control | Drought | Ratio | |||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Linkage group | Position of highest peak (cM) | –log10P | Effect | R2 | Effect | R2 | ||
| CHL | CHL1 | A1 | 24.23 | 4.2 | –0.855 | 5.7 | –0.855 | 5.4 | |
| CHL2 | A6 | 59.11 | 4.5 | –0.885 | 5.9 | –0.885 | 5.6 | ||
| CHL3 | A9 | 77.21 | 8.9 | –1.304 | 14.2 | –1.304 | 13.4 | ||
| CHL4 | A10 | 56.32 | 5.9 | 1.011 | 10.4 | 1.011 | 9.7 | ||
| STC | STC1 | A7 | 96.75 | 3.0 | 6.395 | 1.9 | –9.754 | 7.9 | –1.5 |
| LN | LN1 | A7 | 40.75 | 11.1 | 0.799 | 15.6 | 11.103 | 23.5 | |
| LN2 | A10 | 62.99 | 4.6 | 0.426 | 5.4 | 4.650 | 8.1 | ||
| LA | LA1 | A1 | 70.54 | 6.0 | –3.343 | 15.6 | –0.358 | 0.9 | 9.3 |
| LA2 | A7 | 32.05 | 4.8 | –1.251 | 2.2 | –1.251 | 10.7 | ||
| LA3 | A8 | 85.20 | 5.2 | 1.321 | 2.4 | 1.321 | 11.9 | ||
| LA4 | A9 | 24.28 | 3.4 | 1.865 | 4.8 | –0.394 | 1.1 | –4.7 | |
| LFW | LFW1 | A3 | 42.66 | 3.2 | 0.313 | 7.6 | 0.062 | 3.7 | 5.0 |
| LFW2 | A7 | 105.32 | 3.6 | 0.092 | 0.7 | 0.092 | 8.1 | ||
| LFW3 | A8 | 85.20 | 6.1 | 0.120 | 1.1 | 0.120 | 13.7 | ||
| LDW | LDW1 | A3 | 42.66 | 3.5 | 0.036 | 9.9 | 0.010 | 3.0 | 3.7 |
| LDW2 | A7 | 100.81 | 3.9 | 0.016 | 2.1 | 0.016 | 8.4 | ||
| LDW3 | A8 | 91.33 | 7.3 | 0.023 | 4.0 | 0.023 | 16.5 | ||
| 3,4DW | 3,4DW1 | A5 | 69.71 | 2.8 | 0.011 | 8.7 | 0.003 | 1.3 | 3.4 |
| 3,4DW2 | A7 | 34.89 | 7.4 | –0.011 | 8.5 | –0.011 | 14.9 | ||
| 3,4DW3 | A7 | 100.81 | 3.0 | 0.007 | 3.1 | 0.007 | 5.5 | ||
| DWR | DWR1 | A3 | 120.61 | 3.9 | 0.024 | 6.1 | 0.024 | 4.2 | |
| DWR2 | A4 | 75.90 | 2.9 | 0.020 | 4.4 | 0.020 | 3.0 | ||
| DWR3 | A7 | 40.75 | 18.3 | –0.054 | 31.1 | –0.076 | 42.8 | 1.4 | |
| SDW | SDW1 | A3 | 42.66 | 3.2 | 0.038 | 9.7 | 0.011 | 2.9 | 3.6 |
| SDW2 | A7 | 83.27 | 3.0 | 0.016 | 1.8 | 0.016 | 7.0 | ||
| SDW3 | A8 | 91.33 | 7.0 | 0.025 | 4.2 | 0.025 | 16.3 | ||
| SLA | SLA1 | A5 | 60.82 | 4.6 | –20.3 | 15.0 | 0.201 | 0.0 | –101.4 |
| HL | HL1 | A3 | 94.58 | 6.6 | 0.245 | 11.0 | 0.245 | 12.5 | |
| HL2 | A4 | 54.79 | 3.7 | 0.176 | 5.7 | 0.176 | 6.5 | ||
| HL3 | A6 | 101.21 | 6.6 | 0.265 | 12.9 | 0.265 | 14.7 | ||
| HL4 | A7 | 18.39 | 4.7 | 0.200 | 7.3 | 0.200 | 8.3 | ||
| HDW | HDW1 | A3 | 38.29 | 4.9 | 0.003 | 4.9 | 0.003 | 8.8 | |
| HDW2 | A6 | 62.85 | 4.0 | 0.002 | 3.8 | 0.002 | 6.8 | ||
| HDW3 | A7 | 3.99 | 6.8 | 0.004 | 14.0 | 0.002 | 5.9 | 2.0 | |
| HDW4 | A8 | 33.95 | 4.1 | 0.003 | 6.3 | 0.003 | 11.3 | ||
| RL | RL1 | A5 | 69.71 | 4.1 | 126.3 | 10.6 | 30.215 | 2.7 | 4.2 |
| RL2 | A8 | 21.23 | 3.3 | 120.1 | 9.6 | 32.583 | 3.2 | 3.7 | |
| RL3 | A8 | 86.57 | 5.0 | 62.1 | 2.6 | 62.124 | 11.5 | ||
| RV | RV1 | A5 | 69.71 | 3.2 | 0.141 | 9.6 | 0.056 | 2.4 | 2.5 |
| RV2 | A8 | 86.57 | 5.7 | 0.124 | 7.4 | 0.124 | 11.8 | ||
| RD | RD1 | A3 | 5.92 | 5.6 | –0.010 | 10.8 | –0.010 | 4.6 | |
| RD2 | A5 | 35.17 | 4.8 | –0.008 | 7.8 | –0.008 | 3.3 | ||
| RD3 | A6 | 48.53 | 2.6 | –0.005 | 2.8 | 0.010 | 5.0 | –2.0 | |
| RD4 | A8 | 95.50 | 4.3 | –0.007 | 6.0 | –0.007 | 2.6 | ||
| RL/SDW | RL/SDW1 | A3 | 21.88 | 2.3 | –133.0 | 2.8 | –133.0 | 3.1 | |
| RL/SDW2 | A7 | 18.39 | 6.2 | –236.8 | 8.8 | –236.8 | 10.0 | ||
| RL/SDW3 | A7 | 125.27 | 2.7 | 160.8 | 4.1 | –142.8 | 3.6 | –1.1 | |
| RL/SDW4 | A10 | 62.99 | 4.0 | 182.5 | 5.2 | 182.5 | 5.9 | ||
| RDW | RDW1 | A5 | 69.71 | 3.1 | 0.005 | 4.4 | 0.005 | 6.7 | |
| RDW2 | A8 | 86.57 | 4.9 | 0.006 | 6.7 | 0.006 | 10.3 | ||
| R/S | R/S1 | A7 | 18.39 | 3.6 | –0.014 | 8.2 | –0.014 | 3.7 | |
| R/S2 | A9 | 84.14 | 3.0 | 0.004 | 0.7 | –0.021 | 8.5 | 5.1 | |
| S/SR | S/SR1 | A4 | 90.12 | 3.2 | –0.010 | 6.3 | –0.010 | 4.7 | |
| S/SR2 | A9 | 69.95 | 3.4 | 0.001 | 0.0 | –0.013 | 8.2 | –14.9 | |
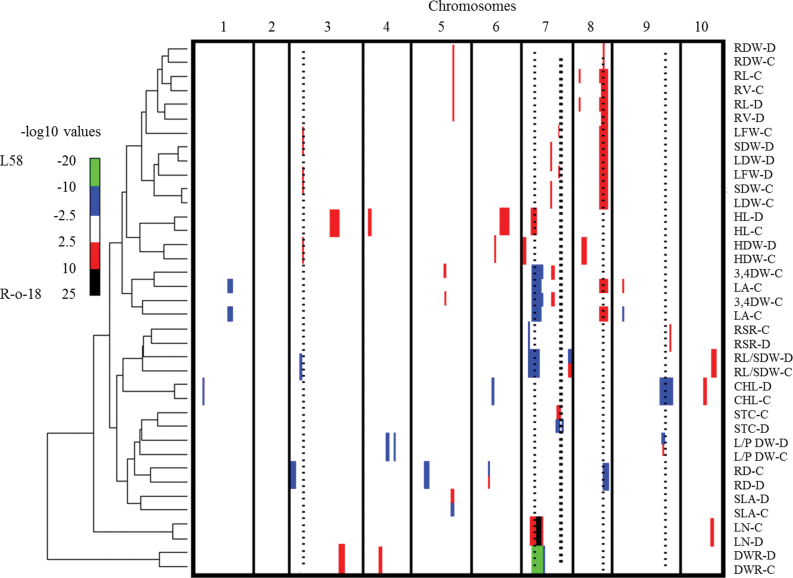
Fig. 2.
A clustered heat map showing the –log10P profiles of the measured traits. Columns indicate the 10 chromosomes in centiMorgans, ascending from left to right; rows indicate individual trait –log10P profiles. A colour scale is used to indicate the QTL significance corresponding to the –log10P score: red and black represent a positive effect on the trait value from the R-o-18 allele; blue and green represent a positive effect on the trait value from the L58 allele. Bar width indicates the significance interval of the QTL. Hierarchical clustering, shown on the left, reflects the correlation between traits based on the QTL profiles. Right dotted line of heavier weight in A7 indicates the QTL with an antagonistic fitness effect; the other five dotted lines refer to QTLs confirmed in the reproducibility experiment. C and D refer to control and drought environments, respectively. Trait abbreviations are given with Table 1.
Stomatal conductance QTL (STC1) and fitness trade offs in the drought environment
The correlation analysis showed that, in the control environment, stomatal conductance was positively correlated with LDW and SDW. On the other hand, in the drought environment, stomatal conductance showed a significant negative correlation with LDW and SDW. These correlations were associated with altering the trait-value-enhancing allele for STC1 from R-o-18 in the control environment to L58 in the drought environment. The trait-value-enhancing alleles for the QTL colocating with STC1 (LFW2, LDW2, 3,4DW2) were R-o-18 in both environments (Table 3, Fig. 2). This means that the R-o-18 alleles for these loci were enhancing fitness both under control and drought conditions, although having contrasting phenotypic effects on stomatal conductance when comparing both conditions.
Mapping QTL underlying plasticity
Seventeen of the mapped QTL showed a significant Q×E effect (Table 3) indicating the loci contributing to phenotypic plasticity between both environments. In addition to the GenStat method to determine these plasticity loci, an alternative method to describe QTL that are affected by the environments was suggested by Tétard-Jones et al. (2011). This uses the differences between the trait-value averages of the lines in the two environments to determine QTL. Using this procedure, 15 plasticity QTL were mapped (Table 4), with nine of them colocating with previously mapped QTL, six of which were found to show Q×E (Table 3). Thus, this analysis detected six new plasticity QTL, which did not exceed the statistical significance levels with the GenStat method.
Table 4.
QTL mapped for phenotypic plasticity in the L58 x R-o-18 RIL populationPlasticity was calculated as described by Tétard-Jones et al. (2011), as the difference in the mean phenotype between different treatments per trait. QTLs are numbered according to chromosome. R 2 is the percentage of total plastic variance explained by each QTL. Effects with positive values represent a positive contribution of the R-o-18 allele to the trait value and those with negative values represent a positive contribution of the L58 allele to the trait value. Highlighted QTL were mapped before using the multienvironment analysis approach (Table 3). Trait abbreviations are given with Table 1.
| Trait | QTL name | Locus | Chromosome | Position (cM) | Effect | –log10P | R 2 |
|---|---|---|---|---|---|---|---|
| STC | STC1 | 903607|9917837 | 7 | 96.8 | 17.22 | 3.8 | 11.0 |
| LA | LA1 | E3835M11 | 1 | 69.0 | –2.69 | 4.6 | 13.3 |
| LDW | LDW4 | E3850M9 | 5 | 69.7 | 0.03 | 3.2 | 9.2 |
| 3,4DW | 3,4DW3 | Ra2A01-A7 | 7 | 83.3 | –0.01 | 2.9 | 8.3 |
| SDW | SDW4 | E3850M9 | 5 | 69.7 | 0.03 | 3.0 | 8.8 |
| SLA | SLA2 | E3749M6 | 1 | 94.1 | 17.26 | 3.0 | 7.4 |
| SLA1 | BrID101239-A5 | 5 | 65.7 | –22.36 | 4.0 | 12.4 | |
| HL | HL5 | 902225|9924661 | 8 | 95.5 | –0.08 | 3.0 | 8.0 |
| RL | RL4 | E3732M5 | 1 | 92.1 | –101.68 | 3.3 | 7.9 |
| RL1 | E3850M9 | 5 | 69.7 | 96.41 | 2.9 | 7.1 | |
| RL3 | E3416M22 | 8 | 91.3 | 112.99 | 4.0 | 9.8 | |
| RV | RV3 | E3732M5 | 1 | 92.1 | –0.12 | 3.0 | 8.1 |
| RD | RD3 | 899015|9918455 | 6 | 43.5 | –0.02 | 3.6 | 9.7 |
| RL/SDW | RL/SDW3 | C7P119 | 7 | 119.0 | 304.08 | 3.4 | 11.8 |
| R/S | R/S2 | BrID10177-A9 | 9 | 68.3 | –0.02 | 2.7 | 7.0 |
Reproducibility
From the 11 QTL clusters that were mapped, four were selected to be tested for reproducibility in a subsequent experiment. The first cluster mapped to A3, including the RD1 and RL/SDW1 QTL for both positively correlated traits, with trait-value-enhancing effects from the L58 alleles. Moreover, LFW1, LDW1, and SDW1, all contributing to shoot biomass, were mapped to the same cluster, with positive alleles coming from R-o-18. The second cluster was mapped to A7—composed of LN1, LA2, 3,4DW2, DWR3, R/S1, and RL/SDW2—all with a positive contribution of the R-o-18 allele except for LN1. This is in line with the negative correlation of leaf number with the other traits. The third cluster, on A8, included eight colocating QTL—LFW3, LDW3, SDW3, LA3, RL3, RVl2, RD4,and RDW2—of which the RD4 L58 allele increased the trait value, while for the other QTL, the R-o-18 allele increased the trait value, in line with the negative correlation of RD with the other traits. The fourth cluster included three QTL—CHL3, R/S2, and S/SR2—mapping to A9. The S/SR ratio showed a negative correlation between control and drought environments and therefore the trait-value-enhancing effect of S/SR2 in the drought environment came from the L58 allele, whereas in the control environment it came from the R-o-18 allele.
In total, 27 lines were selected from the RIL population to properly represent the 16 possible genotypes for all allelic combinations for the four selected QTL clusters. These lines were regrown under similar conditions and rephenotyped (Supplementary Table 1, available at JXB online). A correlation analysis (Table 5) between traits measured in the two control environments and between traits measured in the two drought environments of the full RIL screening and QTL reproducibility experiments showed that all traits were positively correlated in at least one environment, but often both, except for fresh weight, LWC, and root diameter. This indicates that the phenotyping was robust and the detected QTL clusters are reproducible, making them attractive candidates for further gene cloning experiments.
Table 5.
Correlation analysis between the control conditions and the drought conditions of the full RIL screening experiment and the reproducibility experimentHighlighted results refer to significant correlations: dark grey, P < 0.01; light grey, P < 0.05. Trait abbreviations are given with Table 1.
| Trait | Control | Drought |
|---|---|---|
| STC | 0.260 | 0.494 |
| LN | 0.852 | 0.758 |
| LA | 0.504 | 0.220 |
| LFW | 0.122 | –0.027 |
| LDW | 0.270 | 0.258 |
| 3,4DW | 0.474 | 0.239 |
| DWR | 0.296 | 0.128 |
| SDW | 0.266 | 0.259 |
| SLA | 0.496 | 0.159 |
| HL | 0.850 | 0.847 |
| HDW | 0.211 | 0.324 |
| RL | 0.209 | 0.085 |
| RV | –0.002 | 0.292 |
| RD | 0.071 | –0.107 |
| RL/SDW | 0.095 | 0.378 |
| RDW | 0.073 | 0.251 |
| R/S | 0.310 | 0.352 |
| S/SR | 0.397 | 0.476 |
| LWC | 0.105 | 0.088 |
Discussion
The current study was carried out in a greenhouse using pots filled with sand. This type of pot experiment is a reasonable compromise to avoid the difficulty of phenotyping roots in natural field environments and the unnatural conditions present in hydroponics, aeroponics, or agar plates (Tuberosa, 2012). However, aspects of root growth in this pot system would have still been substantially different from field conditions.
Upon screening the RIL population, this work found significant G×E between control and drought environments for stomatal conductance, leaf number, root length, and root diameter. This G×E was reflected in Q×E detected using the MEA approach for these traits, except for leaf number. MEA is more powerful than the traditional single environment analysis in detecting more significant QTL with higher explained variance. An additional advantage is that it allows quantification of Q×E, because it accounts for G×E and tests all detected QTL in all environments and thus shows their effects in each environment (Crossa and Federer, 2012). Q×E occurs if the QTL effects are strongly expressed in one environment but weakly in another, or if the QTL has opposite effects on the same trait in two different environments (Mackay, 2001; MacMillan et al., 2006; Zhang et al., 2010). Examples of the first case are LFW1, LA1, LDW1, 3,4DW1, RL1, RL2, RV1, R/S2, HDW3, and SDW1, while examples of the latter case are found for LA4, SLA1, RD3, and S/SR2. The latter kind of Q×E obstructs the transferability of QTL mapping results from one environment to another (Mackay, 2001), as selection will be in opposite directions in the two environments.
Knowing about the QTL with opposite effects on several traits in different environments, also known as antagonistic pleiotropy, is of great importance in breeding programmes because breeding for one trait might negatively affect other traits (Rose, 1982; Juenger, 2013). The QTL cluster mapped at the bottom of A7 included a stomatal conductance QTL (STC1), which showed signs of antagonistic pleiotropy, with the R-o-18 allele increasing stomatal conductance under control conditions and decreasing it under drought conditions, while having positive effects on biomass under both environments through the colocated LDW3, SDW2, and 3,4DW3 QTL. However, the similar effect on biomass and the contrasting effect on stomatal conductance could also mean these traits are not allelic, but the result of close linkage of two loci. Further analysis should reveal this.
Stomatal conductance showed clear plasticity, decreasing significantly in the drought environment. Such response is generally correlated with reduced photosynthesis but also with reduced water loss as an adaptive response to drought (Chaves et al., 2003; Condon et al., 2004; Tardieu, 2013). Due to the colocation or antagonistic pleiotropy of the shoot biomass QTL with STC1, when comparing both environments, stomatal conductance was negatively correlated with shoot biomass (Table 2B). This reflects an interesting fitness advantage for plants carrying the R-o-18 allele at this QTL cluster, meaning that under drought conditions they show relatively reduced stomatal conductance (contributing to increased drought tolerance) accompanied with relatively increased shoot biomass, compared to plants carrying the L58 allele.
Recently, the plasticity and the evolution of flowering time and water use efficiency (WUE) has been investigated in B. rapa under drought environments (Franks, 2011), and the relationship between circadian rhythm, vegetative, and reproductive traits, and leaf gas exchange with the variation of WUE in different watering regimes has been investigated (Edwards et al., 2012). The negative correlation that this work found for stomatal conductance and shoot biomass under drought was also observed by (Edwards et al., 2012), although this was not significant in their study. It also agrees with the positive correlation between WUE and biomass in the drought environment found by these authors and the colocation of WUE and stomatal conductance QTL mapped in B. rapa grown under warm and long-day conditions (Edwards et al., 2011). Although the preferred targets for crop improvement in marker-assisted breeding are generally constitutively expressed QTL (Bernardo, 2008), this QTL cluster is attractive to select for, even if it is not constitutive in view of the Q×E observed for STC1, because the allele from R-o-18 contributes to increased drought tolerance without having fitness costs due to reducing biomass.
The leaf area response and the underlying QTL in both environments were confirmed by the positive correlation observed between the full RIL screening and QTL reproducibility experiments. Stomatal closure and limited expansion of young leaves under drought have an indirect negative effect on root growth (Chaves et al., 2003; Roycewicz and Malamy, 2012). This was observed by the reduction in root length, concomitant with an increase in root diameter in the drought treatment, corresponding to similar observations reported before for Brassica and other crops (Zhu et al., 2011; Edwards et al., 2012; Poorter et al., 2012). It thus appears that, under drought stress in pots, B. rapa does not invest in longer roots to take up more water, but in thicker roots to act as a water storage buffer.
Under drought, the R/S ratio increased compared to the well-watered conditions. Biomass allocation under limiting environments can be explained by a functional biomass equilibrium when plants allocate more biomass to roots when the factor limiting growth is below ground (e.g. water or nutrient shortage), to enhance the uptake of that limiting factor (Poorter et al., 2012). The correlation of the R/S ratio with drought tolerance has previously also been documented for Arabidopsis and tobacco (Werner et al., 2010), as well as B. rapa (Kage et al., 2004; Edwards et al., 2012).
Of the traits examined, G×E was found for most of them, either in the full RIL screening or the reproducibility experiment (Table 1). With so many traits for which G×E was found, it is not surprising that QTL with Q×E were also found, indicating plasticity for many traits. This work used two ways to detect QTL related to phenotypic plasticity, first using the MEA approach (Table 3) and subsequently using the difference between average values per line when comparing both treatments per lines (Table 4). As previously found by Tétard-Jones et al. (2011), there is considerable overlap between both methods, but the latter method also detects some novel QTL not found previously. This is probably due to the additional statistical power that can be gained by directly using the phenotypic difference values for mapping, meaning that QTL that did not exceed the threshold in the MEA approach will be detected.
Although almost all traits showed a positive correlation between the results from the full RIL screening experiment and the reproducibility experiment, confirming the initial results, this was not the case for leaf fresh weight and root diameter, suggesting a high level of G×E for those traits, or for water content, where a high environmental effect probably prevented mapping a QTL for this trait. The only trait for which no plasticity QTL was found was chlorophyll content (CHL), which was in line with the inability to detect G×E for this trait (Table 1). However, there was genetic variation for CHL, with four detected QTL (Table 3). There is also a difference in CHL between drought and control conditions, which agrees with previous observations for four Brassica species (Ashraf and Mehmood, 1990), but the genotypes appeared to respond similarly to drought exposure by decreasing CHL, explaining the lack of G×E. The four CHL QTL mapped to regions previously identified to contain QTL for chlorophyll a and b content in B. rapa (Ge et al., 2012), with CHL1 colocating with one of the three QTL previously identified for chlorophyll fluorescence (Edwards et al., 2011).
Increasing crop productivity under drought conditions is the ultimate goal for marker-assisted breeding programmes. In that respect, the significant antagonistic effect of relatively reduced stomatal conductance along with relatively higher shoot biomass under drought conditions due to the STC1/shoot biomass locus at the bottom of chromosome A7 is very interesting, as it suggests that selection on reduced water loss during drought, through reduced stomatal transpiration, is expected to have disproportionately little effect on shoot biomass reduction, which is a favourable combination. In addition, this work reported many QTL underlying several morphological and physiological traits, which appeared to be robust and thus provided the first step towards identifying genes governing those traits. The availability of the whole B. rapa genome sequence (Wang et al., 2011) together with possible comparative alignment with the related model species A. thaliana (Schranz et al., 2006) will facilitate fine mapping and cloning of candidate genes underlying the desired QTL. This approach will not only be useful in breeding B. rapa, but also in breeding other closely related species like B. juncea and B. napus (Cheng et al., 2011; Li et al., 2013).
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Phenotypic data of the selected lines for the analysed traits by full RIL screening and QTL reproducibility under control and drought conditions.
Acknowledgements
The Dutch graduate school Experimental Plant Sciences is acknowledged for financially supporting this research. The authors thank Nihal Erol-Öztolan for her help with sowing and harvesting and Unifarm for taking care of the plants in the greenhouse.
References
- Anderson JT, Lee C-R, Rushworth CA, Colautti RI, Mitchell-Olds T. 2013. Genetic trade-offs and conditional neutrality contribute to local adaptation. Molecular Ecology 22, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M, Mehmood S. 1990. Response of four Brassica species to drought stress. Environmental and Experimental Botany 30, 93–100 [Google Scholar]
- Bagheri H, El-Soda M, van Oorschot I, et al. 2012. Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Frontiers in Plant Science 3, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R. 2008. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Science 48, 1649–1664 [Google Scholar]
- Boer MP, Wright D, Feng L, Podlich DW, Luo L, Cooper M, van Eeuwijk FA. 2007. A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177, 1801–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS. 2003. Understanding plant responses to drought-from genes to the whole plant. Functional Plant Biology 30, 239–264 [DOI] [PubMed] [Google Scholar]
- Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. 2011. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biology 11, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis BR, Smith AB, Coombes NE. 2006. On the design of early generation variety trials with correlated data. Journal of Agricultural Biological and Environmental Statistics 11, 381–393 [Google Scholar]
- Crossa J, Federer WT. 2012. Screening experimental designs for quantitative trait loci, association mapping, genotype-by-environment interaction, and other investigations. Frontiers in Physiology 3:156 10.3389/fphys.2012.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CE, Ewers BE, McClung CR, Lou P, Weinig C. 2012. Quantitative variation in water-use efficiency across water regimes and its relationship with circadian, vegetative, reproductive, and leaf gas-exchange traits. Molecular Plant 5, 653–668 [DOI] [PubMed] [Google Scholar]
- Edwards CE, Ewers BE, Williams DG, Xie Q, Lou P, Xu X, McClung CR, Weinig C. 2011. The genetic architecture of ecophysiological and circadian traits in Brassica rapa . Genetics 189, 375–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ. 2011. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa . New Phytologist 190, 249–257 [DOI] [PubMed] [Google Scholar]
- Ge Y, Wang T, Wang N, Wang Z, Liang C, Ramchiary N, Choi S-R, Lim YP, Piao ZY. 2012. Genetic mapping and localization of quantitative trait loci for chlorophyll content in Chinese cabbage (Brassica rapa ssp. pekinensis). Scientia Horticulturae 147, 42–48 [Google Scholar]
- Juenger TE. 2013. Natural variation and genetic constraints on drought tolerance. Current Opinion in Plant Biology 16, 274–281 [DOI] [PubMed] [Google Scholar]
- Kage H, Kochler M, Stützel H. 2004. Root growth and dry matter partitioning of cauliflower under drought stress conditions: measurement and simulation. European Journal of Agronomy 20, 379–394 [Google Scholar]
- Levitt J. 1972. Responses of plants to environmental stresses. New York: Academic Press [Google Scholar]
- Li X, Ramchiary N, Dhandapani V, Choi SR, Hur Y, Nou I-S, Yoon MK, Lim YP. 2013. Quantitative trait loci mapping in Brassica rapa revealed the structural and functional conservation of genetic loci governing morphological and yield component traits in the A, B, and C subgenomes of Brassica species. DNA Research 20, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC. 2001. The genetic architecture of quantitative traits Annual Review of Genetics 35, 303–339 [DOI] [PubMed] [Google Scholar]
- MacMillan K, Emrich K, Piepho HP, Mullins C, Price A. 2006. Assessing the importance of genotype × environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. Theoretical and Applied Genetics 113, 953–964 [DOI] [PubMed] [Google Scholar]
- Malosetti M, Voltas J, Romagosa I, Ullrich SE, van Eeuwijk FA. 2004. Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica 137, 139–145 [Google Scholar]
- Poorter H, Niklas KJ, Reich PB, Oleksyn J, Poot P, Mommer L. 2012. Biomass allocation to leaves, stems and roots: meta-analyses of interspecific variation and environmental control. New Phytologist 193, 30–50 [DOI] [PubMed] [Google Scholar]
- Rose MR. 1982. Antagonistic pleiotropy, dominance, and genetic variation. Heredity 48, 63–78 [Google Scholar]
- Roycewicz P, Malamy JE. 2012. Dissecting the effects of nitrate, sucrose and osmotic potential on Arabidopsis root and shoot system growth in laboratory assays. Philosophical Transactions of the Royal Society B 367, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting CD. 1986. The evolution of phenotypic plasticity in plants. Annual Review of Ecology and Systematics 17, 667–693 [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. 2006. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Science 11, 535–542 [DOI] [PubMed] [Google Scholar]
- Smith S, De Smet I. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Philosophical Transactions of the Royal Society B 367, 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhwinder S, Hernandez MV, Crossa J, Singh PK, Bains NS, Singh K, Sharma I. 2012. Multi-trait and multi-environment QTL analyses for resistance to wheat diseases. PLoS One 7, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F. 2013. Plant response to environmental conditions: assessing potential production, water demand and negative effects of water deficit. Frontiers in Physiology 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétard-Jones C, Kertesz MA, Preziosi RF. 2011. Quantitative trait loci mapping of phenotypic plasticity and genotype–environment interactions in plant and insect performance. Philosophical Transactions of the Royal Society B 366, 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa R. 2012. Phenotyping for drought tolerance of crops in the genomics era. Frontiers in Physiology 3, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, Lande R. 1985. Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa . Nature Genetics 43, 1035–1039 [DOI] [PubMed] [Google Scholar]
- Welham SJ, Gogel BJ, Smith AB, Thompson R, Cullis BR. 2010. A comparison of analysis methods for late-stage variety evaluation trials. Australian and New Zealand Journal of Statistics 52, 125–149 [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. 2010. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. The Plant Cell 22, 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Y-X, Wang Y, et al. 2010. Stability of QTL across environments and QTL-by-environment interactions for plant and ear height in maize. Agricultural Sciences in China 9, 1400–1412 [Google Scholar]
- Zhu JM, Ingram PA, Benfey PN, Elich T. 2011. From lab to field, new approaches to phenotyping root system architecture. Current Opinion in Plant Biology 14, 310–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.