Abstract
Proper regulation of cell proliferation, cell apoptosis, and cell death are vital for the development and survival of living organisms. Failure or dysfunction of any of these processes can have devastating effects, including cancer. The Hippo pathway, first discovered in Drosophila, has been found to be a major growth-regulatory signaling pathway that controls these crucial processes and has been implicated in cell-progress regulation and organ size determination. Abnormal regulation of this pathway has been found in several cancer types. However, the mechanisms that regulate the pathway and its core members yet have to be elucidated. One of the main core components of this pathway is LATS1, a serine/threonine kinase. Therefore, understanding how LATS1 activity is regulated is expected to shed light on new mechanisms that regulate the Hippo pathway. In the current work, we identified several potential LATS1 regulators and proved that NEDD4 E3 ubiquitin ligase controls LATS1 stability. We demonstrate that NEDD4 directly interacts with LATS1, leading to ubiquitination and decreased levels of LATS1 and, thus, increased YAP localization in the nucleus, which subsequently increases the transcriptional activity of YAP. As such, we show that NEDD4 acts as an additional regulator of the Hippo pathway on the protein level via interactions between WW domain-containing and PPxY motif-containing proteins. These findings might be applied in the development of new therapeutic approaches through the activation of LATS1.
Keywords: NEDD4, LATS1, Hippo pathway, WW domain, protein-protein interaction
Introduction
The Hippo pathway is a highly conserved pathway that regulates organ size by directing cell proliferation and apoptosis in addition to mediating cell growth, cell fate, and stem cell identity. The Hippo pathway is regulated by cellular architecture and environmental properties and may serve as a sensor of tissue structure and mechanical tension as well as regulating cytoskeletal dynamics.1,2
The pathway is activated by high cell density, which subsequently suppresses cell proliferation and may induce apoptosis.3,4 In mammals, when the pathway is activated, a core kinase cascade is activated as follows: MST1/2 kinase interacts with and phosphorylates WW45, an adaptor protein. Together, this complex phosphorylates and activates LATS1/2, which, together with its co-factor MOB1, phosphorylates YAP (or TAZ, depending on cell context). Once phosphorylated, YAP is sequestered or degraded in the cytoplasm.3,5 Both YAP and TAZ, most downstream effectors of the pathway, function as transcription co-activators to inhibit apoptosis and promote cell proliferation.4,6
LATS1 is a tumor suppressor serine–threonine kinase that contains 2 PPxY motifs that are able to bind with high affinity to WW domain-containing proteins.7-9 In addition to its role within the Hippo pathway, LATS1 also plays a role in regulating the cell cycle and apoptosis. LATS1 blocks the G2/M transition during the cell cycle by modulating levels of Cyclin A and B, thereby, arresting cell proliferation and inhibiting growth.7 Additionally, LATS1 is localized to the centrosome and involved in spindle formation during mitosis. Dysfunction of this important process can lead to genetic instability, irregular cellular segregation, and chromosomal abnormalities. Moreover, LATS1 was found to induce apoptosis and, in certain cell lines, upregulate pro-apoptotic genes such as p53 and Bax in response to extensive DNA damage, UV irradiation, chemotherapeutics, oncogenic activation, and growth factor withdrawal. These functions require the kinase activity of the protein. Thus, LATS1 serves as a tumor suppressor through several different mechanisms that negatively regulate tumor development.9-11
Downregulation of LATS1/2 has been shown in human sarcomas, ovarian sarcomas, breast cancers, astrocytomas, retinoblastomas, and acute lymphoblastic leukemia.12-14 Several oncogenic micro-RNAs have also been implicated in the regulation of LATS1/2 and are overexpressed in some types of cancer, such as AGS human gastric15 and testicular cancers.11 Recently, it was reported that LATS1 is regulated at the protein level through ubiquitin-mediated degradation.16-18 Thus, it could be speculated that lower levels of LATS1/2 directly increase the oncogenic nuclear activity of YAP, which might be responsible for the advancement of many of the abovementioned cancers.
NEDD4 is an E3 ubiquitin ligase with a HECT catalytic domain and 4 WW domains and is involved in ubiquitin-dependent protein degradation, protein trafficking, and nuclear localization of multiple proteins. It has a strong affinity for proline-rich proteins, and many of its substrates carry PY (proline-tyrosine, PPxY) motifs. It is part of the NEDD4-like family of E3 ubiquitin ligase proteins, which also include WWP1/2, Smurf1/2, NEDDL1/2, NEDD4L, and ITCH.19,20 NEDD4 was first discovered as a developmentally regulated gene in the mouse central nervous system and was found to target the epithelial sodium channel. Yet many more substrates for this ligase were subsequently discovered.21 NEDD4 is involved in several different pathways and regulates many important proteins, of which several are linked to cancer development.22-26 For example, overexpression of NEDD4 has been linked to cancers of the prostate, bladder, and stomach.19 In the current work, we present additional evidence of the pro-oncogenic function of NEDD4. We identified NEDD4 as a negative regulator of the Hippo tumor suppressor pathway via its targeting one of the pathway’s main tumor suppressor core kinases, LATS1.
Results
LATS1 is a target of WW domain interaction
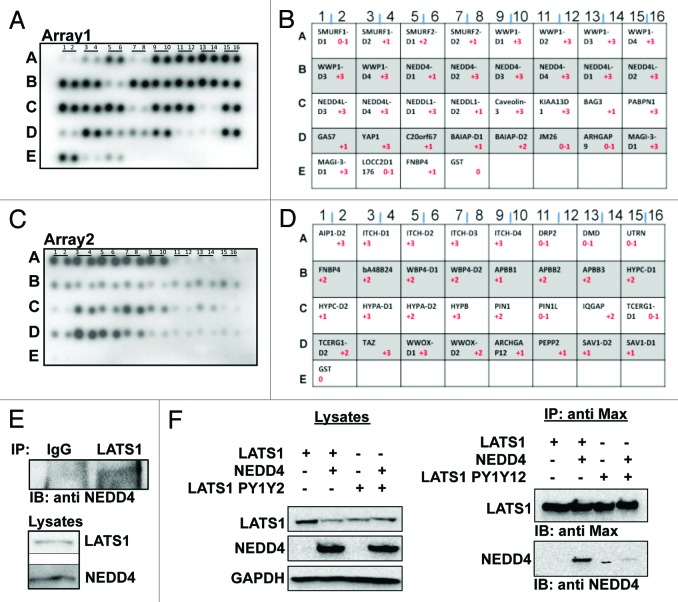
We recently reported that the E3-ubiquitin ligase ITCH physically and functionally regulate LATS1 protein levels and function.18 We therefore assumed that other WW domain-containing proteins might regulate LATS1 function, thereby controlling the Hippo pathway. Using a commercial WW domain peptide array, we screened for WW domain-containing proteins that are able to interact with LATS1. As shown in Figure 1, we detected YAP, TAZ, and ITCH as known, previously reported partners of LATS1 (Fig. 1A–D). Interestingly, this array showed several new WW domain-containing proteins with specific interaction with LATS1 (Fig. 1). For example, WW domains 2, 3, and 4 of NEDD4 bound LATS1 with high affinity (Fig. 1A, row B, lines 7–12). By contrast, the WW1 domain of NEDD4 failed to interact with LATS1 (Fig. 1A, row B, lines 5–6), suggesting a differential binding of NEDD4’s WW domains with LATS1. Similarly, WW domains of WWP1 interacted with high affinity to LATS1 (Fig. 1A, row A, lines 9–16). By contrast, the WW domains of SMURF1/2, members of the NEDD4-like family proteins, did not show high-affinity interactions with LATS1 (Fig. 1A, row A, lines 1–8). In another example, the WW1 domain of WWOX (Fig. 1C, row D, lines 5 and 6), the main interacting WW domain of WWOX,27,28 interacted with higher affinity with LATS1 as compared with its WW2 domain (Fig. 1C, row D, lines 7 and 8). These findings further indicate that LATS1, a key component of the Hippo pathway, is a target of specific WW domain interactions, and that these interactions might regulate the Hippo pathway.
Figure 1. Physical interaction between LATS1 and NEDD4. (A and C), binding pattern of different WW domain containing proteins to LATS1 using commercial WW domain arrays (Panomics). B&D the key for protein WW domains imprinted on WW domain arrays 1 and 2, respectively. (E) endogenous LATS1-NEDD4 interaction. HEK293T cells lysates were immunoprecipitated (IP) with anti-LATS1 and analyzed by immunoblotting (IB) using anti-NEDD4. Cells were pretreated with MG132 for 4 h. Anti-IgG was used as a negative control. (F) NEDD4 forms a complex with LATS1 in vivo. HEK293T cells were cotransfected with either wild-type or mutant Max-LATS1 and NEDD4. Cell lysates were immunoprecipitated with anti-MAX antibodies. The immunoprecipitates were analyzed by immunoblotting as indicated. Lysates were also run and blotted to ensure successful transfection of the cells.
NEDD4 interacts with LATS1 via its PPxY motif
Since LATS1 protein levels seem to be tightly regulated by E3 ubiquitin ligases,16-18 we aimed to test whether the E3 ubiquitin ligase NEDD4 is also capable of mediating LATS1 degradation and modulating the Hippo pathway signaling. To confirm the physical interaction between NEDD4 and LATS1, we performed an immunoprecipitation (IP) assay between the endogenous proteins using HEK293T cells. Cell lysates were immunoprecipitated with polyclonal LATS1 antibody. As a control, we used rabbit anti-IgG. Immunoblotting with anti-NEDD4 antibody revealed endogenous physical interaction between LATS1 and NEDD4 (Fig. 1E). To further confirm that NEDD4 does indeed bind to LATS1, we did an immunoprecipitation assay between the exogenous proteins. HEK293T cells were transfected with Max-LATS1 and NEDD4. After 24 h, cells were lysed and immunoprecipitated with an anti-Max antibody. Indeed we found that NEDD4 co-immunoprecipitates with LATS1 (Fig. 1F).
To map the interaction between LATS1 and NEDD4, we repeated the exogenous interaction between the 2 proteins but using a LATS1 plasmid with both PPxY motifs mutated (LATS1PY1PY2). As shown in Figure 1F, the interaction between LATS1 and NEDD4 was abrogated by PPxY domain mutations, indicating that the NEDD4-LATS1 interaction occurs via the PPxY motifs on LATS1. Taken together, this data indicates that NEDD4, via its WW domains (Fig. 1A), interacts with the PY motifs of LATS1 (Fig. 1F).
NEDD4 ubiquitinates LATS1
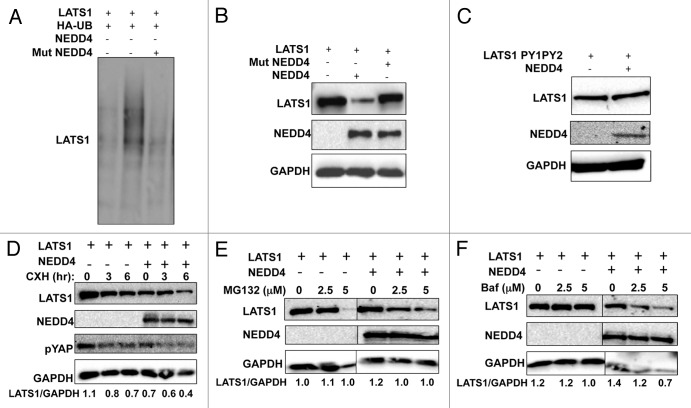
To analyze the functional significance of the interaction between LATS1 and NEDD4, the role of NEDD4 in ubiquitination of LATS1 was analyzed. Extracts of HEK293T cells transfected with plasmids expressing HA-UB, Max-LATS1, and NEDD4, or the catalytically inactive NEDD4 mutant were subjected to immunoprecipitation followed by immunoblotting. Expression of NEDD4 increased the ubiquitination of LATS1 (Fig. 2A lane 2), unlike the inactive mutant of NEDD4 (Fig. 2A lane 3), which was still capable of binding LATS1 (data not shown).
Figure 2. NEDD4 induces ubiquitination and affects the steady-state and half-life of LATS1 protein. (A) NEDD4 ubiquitinates LATS1 in vivo. HEK293T cells were transfected with the indicated plasmids. After 24 h, cells were treated with 20 μmol/L MG132 for 4 h. Lysates were prepared (B) and IP with anti-Max and detected with anti-HA antibodies (UB). (B and C) LATS1 steady-state stability is controlled by NEDD4. HEK293T cells were transiently transfected with Max-LATS1 or LATS1PY1PY2 (C) and NEDD4. After 24 h, cells were lysed and blotted as indicated. (D) ITCH reduces the half-life of LATS1. HEK293 cells were cotransfected with the indicated plasmids. After 24 h, cells were treated with 20 μg/mL cycloheximide (CHX) at the indicated time points and analyzed as shown in the figure. (E and F) LATS1 stability is controlled by the proteasomal pathway. HEK293T cells were transiently transfected with Max-LATS1 and NEDD4. After 24 h cells were treated with MG-132 (E), or bafilomycin [BAF] (F) as indicated for 4 additional hours. Equal amounts of total lysates were blotted as indicated. GAPDH, glyceraldehyde-3-phosphate dehydrogenase was always used as a protein loading control. Numbers under the blots represent the relative expression of LATS1 protein after correction with the loading control GAPDH expression levels.
NEDD4 regulates the stability and half-life of LATS1 and modulates YAP phosphorylation
To determine whether NEDD4-mediated ubiquitination of LATS1 promotes LATS1 degradation, we measured steady-state levels of LATS1 in the presence and absence of NEDD4. Expression of wild-type, but not mutant NEDD4, decreased the protein level of wild-type LATS1 (Fig. 2B) but not LATS1PY1,2 (Fig. 2C).
To prove that the NEDD4-dependent reduction in LATS1 protein was a result of increased degradation, we analyzed the half-life of LATS1 in the presence or absence of NEDD4 using the protein synthesis inhibitor, cycloheximide. Expression of NEDD4 led to a decrease in the protein level of LATS1 (Fig. 2D).
Since LATS1 interacts with and phosphorylates YAP (the most downstream effector of the Hippo pathway), we next determined whether this NEDD4-dependent degradation of LATS1 affects YAP phosphorylation using an antibody that recognizes the phosphorylated serine 127 (S127) on YAP. Indeed, our results demonstrate that decreased levels of LATS1, brought about by NEDD4 overexpression, is accompanied by decreased phospho-YAP levels (Fig. 2D). These results suggest that NEDD4-mediated degradation of LATS1 might affect Hippo signaling.
Because most cellular protein degradation is mediated by the proteasomal pathway, we treated HEK293T cells expressing Max-LATS1 and NEDD4 with increasing amounts of the proteasome inhibitor MG-132 and determined the protein level of LATS1 by western blot. As shown in Figure 2E, treatment with MG132 led to increased protein levels of Max-LATS1 in a dose-dependent manner, whereas treatment with the lysosomal inhibitor, bafilomycin, had no effect on LATS1 levels (Fig. 2F). Altogether, these results indicate that NEDD4 promotes the ubiquitin-dependent proteasomal degradation of LATS1.
NEDD4-mediated degradation of LATS1 leads to accumulation of nuclear YAP
Since YAP localization has been shown to be associated with LATS1-mediated phosphorylation of S127, we next determined whether NEDD4 might affect YAP subcellular localization. GFP-tagged LATS1 (EGFP-LATS1), pDsRed-YAP2 (Red-YAP2), and NEDD4 or mutant NEDD4 were coexpressed in HeLa cells and subcellular localization of NEDD4, LATS1, and YAP was monitored using confocal microscopy (Fig. 3). Co-expression of EGFP-LATS1 and Red-YAP2 showed diffuse and mostly cytoplasmic YAP2 (Fig. 3B). In contrast, coexpression of NEDD4 enhances the localization of LATS1 into cytoplasmic vesicles and the nuclear localization of YAP2 (Fig. 3C).
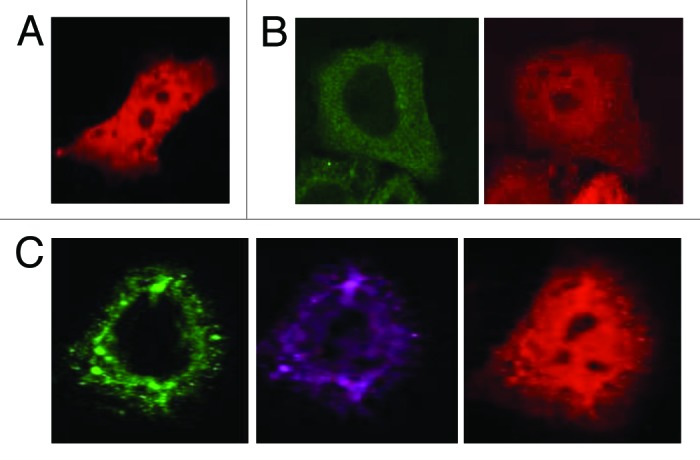
Figure 3. NEDD4 expression is associated with nuclear YAP. HeLa cells were cotransfected with EGFP-LATS1 with DsRed-YAP in the presence or absence of NEDD4. The localization of green fluorescent protein (GFP), Red-tagged, and NEDD4 was visualized by confocal microscopy using 60× magnification.
NEDD4 increases YAP transcriptional transactivation function
YAP functions as a co-activator of the TEAD transcription factors.3,4 Since our data suggest that NEDD4 overexpression may enhance YAP nuclear localization (Fig. 3), we decided to investigate if NEDD4 can affect YAP transactivation function of TEAD. To this end, we transfected a TEAD4-LUC reporter together with YAP2 in HEK293T cells in the presence or absence of NEDD4. Upon overexpressing YAP2 with TEAD4 and TEAD4-LUC reporter, the reporter activity was induced by 20-fold (Fig. 4). In contrast, coexpression of YAP with LATS1 reduced TEAD transactivation to 11-fold. Interestingly, this attenuation was rescued when NEDD4 was co-expressed with LATS1 and YAP. To further confirm the specificity in this assay, we used a mutant NEDD4 that was unable to degrade LATS1, and a mutant LATS1 (PY1PY2) that was unable to interact with NEDD4 as controls. Upon using the mutant forms of LATS1, the reporter activity was not affected as we observed when using wild-type LATS1. In contrast, when we used the mutant form of NEDD4, the reporter activity was significantly attenuated and was comparable to what was obtained upon overexpressing wild type LATS1. These results confirm that NEDD4 increases YAP transcriptional co-activation, at least in part, by downregulating LATS1 levels.
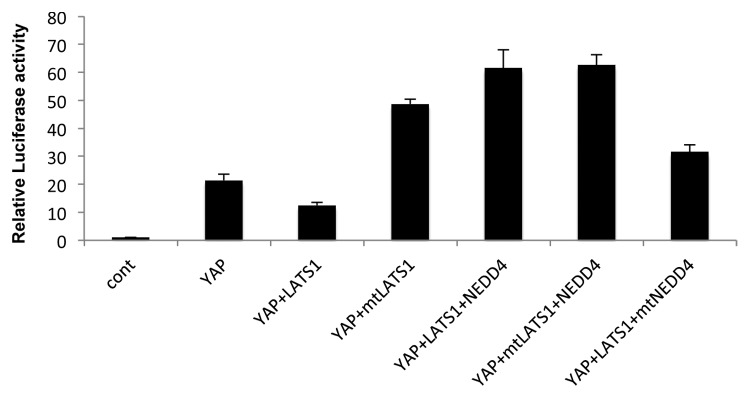
Figure 4. NEDD4 enhances transcriptional co-activation function of YAP. HEK293T were transfected with the indicated plasmids in addition to TEAD response elements cloned in GAL4 plasmid, TEAD transcription factor, and Renilla plasmid as an internal control for transfection efficiency. Twenty-four hours later cells were lysed and relative luciferase activity was measured. Error bars represent standard deviation; P < 0.05.
Discussion
The Hippo pathway is an emerging signaling cascade that plays pivotal roles in regulating organ size, stem cell pluripotency, and tumorigenesis.4,14,29 Protein–protein interactions are a major mediator of the Hippo pathway regulation and activation.5,6 While there are several modular domains present among the various players within the pathway, the WW domain clearly stands out. Indeed, core cascade proteins, upstream regulators, as well as downstream effectors of the pathway were shown to modify the pathway outcomes by involvement of WW domain interactions.18,30-33 To further identify the role of other WW domain containing proteins in regulating the pathway, we searched for LATS1-binding partners using WW domain peptide arrays, since LATS1 is a master regulator of the Hippo pathway. Previously, we identified the E3 ubiquitin ligase ITCH as a negative regulator of the Hippo pathway by interacting with and degrading LATS1. We further published that due to this ITCH-dependent reduction in LATS1 levels, YAP accumulates in the nucleus, leading to enhanced transcriptional co-activation of YAP target genes, enhanced proliferation, survival, tumorigenicity, and epithelial-to-mesenchymal transition.18 In the current work, we found and identified NEDD4 as a novel negative regulator of LATS1. We have demonstrated that NEDD4 physically interacts with LATS1 through binding of the WW-PPxY domains, leading to ubiquitination and degradation of LATS1 in the proteasome. We further showed that overexpression of NEDD4 is associated with increased YAP translocation into the nucleus and the activation of YAP transcriptional transactivation function.
It seems that ITCH and NEDD4 are not the only E3 ligases that control LATS1 stability. Our WW peptide arrays suggest that additional E3 ligases appear to regulate LATS1 function including WWP1, NEDD4L, and NEDD4L1. Consistent with these observations, Yeung et al. have recently demonstrated that the WWP1 E3 ligase is capable of binding LATS1 and mediating its degradation.16 This WWP1-mediated LATS1 degradation was shown to affect cell proliferation in breast cancer cells. Although in this article,16 the authors were able to show that, like WWP1, NEDD4 overexpression reduces LATS1 protein levels, they failed to show that NEDD4 knockdown by siRNA is able to increase the endogenous levels of LATS1. This could likely be related to the cell type that was used, as the endogenous levels of both NEDD4 and LATS1 are much higher than those of WWP1 and ITCH, which might make it difficult to sufficiently deplete NEDD4, or to increase the levels of LATS1. Nevertheless, our data clearly demonstrate that NEDD4 is capable of modulating LATS1 function in vivo.
Although our data suggest that NEDD4 might have an oncogenic function by deregulating the Hippo tumor suppressor pathway, NEDD4 function seems to be complex in this respect. On one hand, it has been shown that NEDD4 negatively regulates the function of different tumor suppressor genes, such as the tumor suppressor PTEN and the anti-angiogenic protein Thrombospondin-1, through which NEDD4 enhances tumorigenesis and tumor progression.22,23,25 On the other hand, NEDD4 negatively regulates the oncogenic IGF-R signaling pathway19,34 as well as the pro-angiogenic factor, VEGF,19,26 and thus suppresses tumor growth and progression. Since NEDD4 regulates the stability of several proteins, we cannot exclude the possibility that other proteins could also contribute to the phenotypes observed in our study.
Previous studies have showed that LATS1 suppresses tumor cell growth by interacting with 2 WW domain proteins, YAP and TAZ, via its PY motifs.4,35 Our findings shed light on other WW domain proteins that might bind LATS1, including NEDD4 and WWOX.36,37 It is therefore possible that under physiological conditions, expression of ITCH, NEDD4, and WWP1 may compete with YAP and TAZ for LATS1 binding to control the levels of LATS1 tumor suppressor. Developing strategies that specifically target these E3 ligases, including NEDD4, or disrupt their interaction with LATS1 in cancer cells to activate LATS1 may be a useful approach for successful cancer therapy.
In conclusion, our study has identified the NEDD4 E3 ligase as a novel negative regulator of LATS1 tumor suppressor stability. Further characterization of the functional interactions and examination of the correlations in clinical samples will provide useful information for future targeting of the NEDD4-LATS1 interaction in cancer intervention.
Materials and Methods
Cell culture and transient transfection
HEK293T cells were grown in DMEM, supplemented with 10% FBS (Gibco), glutamine, and penicillin/streptomycin (Beit-Haemek). Cells were routinely authenticated, and cell aliquots from early passages were used. Transient transfections were achieved using Mirus TransLTi (Mirus Bio LLC).
WW domain arrays
WW Domain Array Kit from Panomics (Panomics, Inc) was used according to manufacturer instructions with slight modifications. In brief, HEK293T cells (10 cm plates) were transiently transfected with HA-LATS1. Forty-eight hours post-transfection, lysates of plates were collected and incubated with pre-washed and blocked array membranes over night at 4 °C with slight shaking. Membranes were washed 3 times with 1× wash buffer for 10 min/each at room temperature. Membranes were then blotted with anti-HA HRP antibody (Roche Applied Science) and detected with ECL.
Immunoprecipitation and immunoblot analysis
Four hours prior to cell lysis, cells were treated with 20 μmol/L of MG132 (Sigma Aldrich). Cells were lysed by using Nonidet P-40 lysis buffer containing 50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 10% glycerol, 0.5% Nonidet P-40, and protease inhibitors. Lysates were precleared with mouse IgG, immunoprecipitations were performed in the same buffer, and lysates were washed 4 times with the same buffer containing 0.1% Nonidet P-40. Western blotting was conducted under standard conditions. Antibodies used were monoclonal anti-Omni (sc-7270, Sanat Cruz), polyclonal anti-NEDD4 (sc-25508, Santa Cruz), polyclonal anti-phospho-YAPS127 (#4911S, Cell Signaling), and monoclonal anti-GAPDH (CB-1001, Calbiochem).
In vivo ubiquitination assay
HEK293 cells were cotransfected with HA-UB, Max-LATS1 with or without NEDD4. After 24 h, cells were treated with MG-132 (20 μmol/L, Sigma) for 4 h. Lysates were immunoprecipitated using anti-Max antibody, washed 4 times, and immunoblotted with anti-HA-HRP (3F10, Roche Applied Science).
Measurement of steady-state and half-life of LATS1 protein level
HEK293T cells were transfected with Max-LATS1 with or without NEDD4. Twenty-four hours posttransfection, cells were lysed or treated with the protein synthesis inhibitor cycloheximide (100 μg/mL, Sigma) for 3 and 6 h. Cell lysates were subjected to immunoblotting.
Immunofluorescence
Cells were seeded on round slide coverslips in 12-well plates. Twenty-four hours later, cells were transfected with the expression plasmids. Twenty-four hours posttransfection, cells were fixed in 3.7% PBS-buffered formaldehyde, permeabilized with 0.05% Triton X-100 at room temperature. Cells were then incubated for 1 h in 10% goat serum (Invitrogen), with primary antibody for 1 h and with secondary antibody. Alexa Fluor 647 goat anti-rabbit IgG (#A21244, Molecular Probes) was used to detect NEDD4. Cells were examined by confocal microscopy (Olympus) under 60× magnification.
Luciferase assay
HEK293 cells seeded in 12-well plates were cotransfected with the relevant plasmids together with TEAD4, TEAD-LUC reporter, and Renilla luciferase as an internal control. Cells were collected 24 h later, and Firefly and Renilla luciferase activities were assayed with Dual-Luciferase Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. All experiments were done at least thrice.
Acknowledgments
Authors would like to thank all members of the Aqeilan lab for fruitful discussion. The research work was supported by funds from the Israel Science Foundation (ISF# 12-0542) to RI Aqeilan.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26672
References
- 1.Schroeder MC, Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol. 2012;23:803–11. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Artemenko Y, Devreotes PN. Hippo on the move: tumor suppressor regulates adhesion and migration. Cell Cycle. 2013;12:535–6. doi: 10.4161/cc.23668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: from development to cancer and beyond. J Biochem. 2011;149:361–79. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 4.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu AM, Wong KF, Jiang X, Qiao Y, Luk JM. Regulators of mammalian Hippo pathway in cancer. Biochim Biophys Acta. 2012;1826:357–64. doi: 10.1016/j.bbcan.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–33. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2011;2:e172. doi: 10.1038/cddis.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 10.Xia H, Qi H, Li Y, Pei J, Barton J, Blackstad M, Xu T, Tao W. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21:1233–41. doi: 10.1038/sj.onc.1205174. [DOI] [PubMed] [Google Scholar]
- 11.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 12.Hisaoka M, Tanaka A, Hashimoto H. Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab Invest. 2002;82:1427–35. doi: 10.1097/01.LAB.0000032381.68634.CA. [DOI] [PubMed] [Google Scholar]
- 13.Chakraborty S, Khare S, Dorairaj SK, Prabhakaran VC, Prakash DR, Kumar A. Identification of genes associated with tumorigenesis of retinoblastoma by microarray analysis. Genomics. 2007;90:344–53. doi: 10.1016/j.ygeno.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-L A, Kenney AM. The Hippo in the room: a new look at a key pathway in cell growth and transformation. Cell Cycle. 2010;9:2292–9. doi: 10.4161/cc.9.12.11919. [DOI] [PubMed] [Google Scholar]
- 15.Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS, Lee JH, Koo KH, Park JW, Kim KS. miR-372 regulates cell cycle and apoptosis of ags human gastric cancer cell line through direct regulation of LATS2. Mol Cells. 2009;28:521–7. doi: 10.1007/s10059-009-0158-0. [DOI] [PubMed] [Google Scholar]
- 16.Yeung B, Ho KC, Yang X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS One. 2013;8:e61027. doi: 10.1371/journal.pone.0061027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho KC, Zhou Z, She YM, Chun A, Cyr TD, Yang X. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability [corrected] Proc Natl Acad Sci U S A. 2011;108:4870–5. doi: 10.1073/pnas.1101273108. [corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71:2010–20. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- 20.Harvey KF, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–9. doi: 10.1016/S0962-8924(99)01541-X. [DOI] [PubMed] [Google Scholar]
- 21.Yang B, Kumar S. Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 2010;17:68–77. doi: 10.1038/cdd.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, Stambolic V, Rotin D. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A. 2008;105:8585–90. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SS, Yoo NJ, Jeong EG, Kim MS, Lee SH. Expression of NEDD4-1, a PTEN regulator, in gastric and colorectal carcinomas. APMIS. 2008;116:779–84. doi: 10.1111/j.1600-0463.2008.00999.x. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Xu LL, Masuda K, Raymundo E, McLeod DG, Dobi A, Srivastava S. A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J Biol Chem. 2008;283:28988–95. doi: 10.1074/jbc.M710528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fouladkou F, Lu C, Jiang C, Zhou L, She Y, Walls JR, Kawabe H, Brose N, Henkelman RM, Huang A, et al. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J Biol Chem. 2010;285:6770–80. doi: 10.1074/jbc.M109.082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murdaca J, Treins C, Monthouël-Kartmann MN, Pontier-Bres R, Kumar S, Van Obberghen E, Giorgetti-Peraldi S. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J Biol Chem. 2004;279:26754–61. doi: 10.1074/jbc.M311802200. [DOI] [PubMed] [Google Scholar]
- 27.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. 2004;101:4401–6. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem. 2009;108:737–45. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 29.Yu FX, Mo JS, Guan KL. Upstream regulators of the Hippo pathway. Cell Cycle. 2012;11:4097–8. doi: 10.4161/cc.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CY, Lv X, Li T, Xu Y, Zhou X, Zhao S, Xiong Y, Lei QY, Guan KL. PP1 cooperates with ASPP2 to dephosphorylate and activate TAZ. J Biol Chem. 2011;286:5558–66. doi: 10.1074/jbc.M110.194019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF, Hong W. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–10. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao L, Chen Y, Ji M, Dong J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem. 2011;286:7788–96. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 36.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65:6764–72. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 37.Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6:249–59. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]