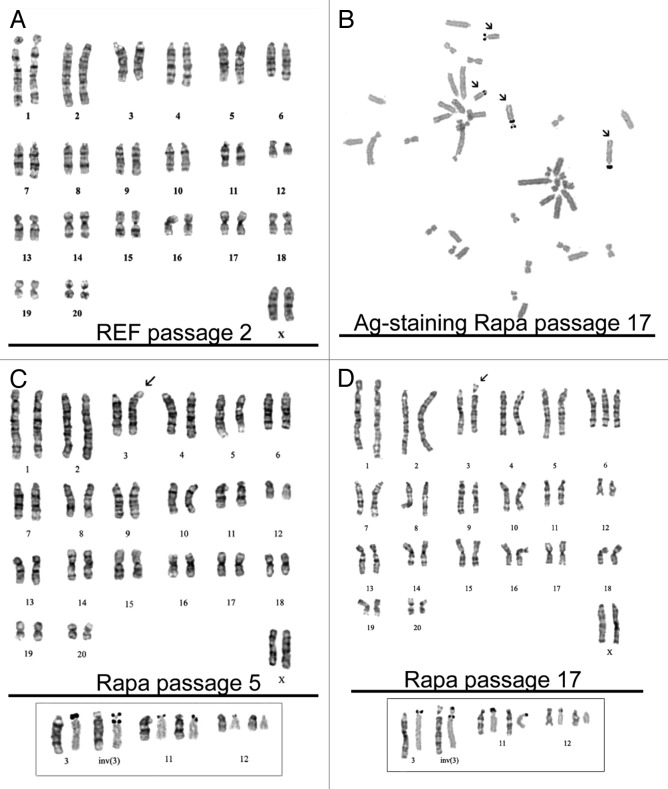
Figure 7. Karyotype analysis of rapamycin-derived cells. (A) REF cells (passage 2) have normal karyotype with a modal chromosome number 42. (B) Chromosomes were stained with silver nitrate (AgNO3) to identify transcriptionally active nucleolar organizers (arrows) for confirmation of the pericentric inversion without loss of nuclear organizers. The insert below shows chromosome pairs with nucleolar organizers: the left chromosome in the pair: G-banding, the right chromosome in the pair: Ag-positive staining. The homolog of chromosome 12 is Ag-negative. (C) Rapamycin-derived cells (passage 5). Modal karyotype: 42 chromosomes, XX, inv (3) (pter → p12:: q21 → p12:: q21 → qter). The arrow shows the pericentric inversion in chromosome 3. (D) Rapamycin-derived cells (passage 17). Modal karyotype: 43XX, inv (3) (pter → p12:: q21 → p12:: q21 → qter). The arrow shows the pericentric inversion in chromosome 3.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.