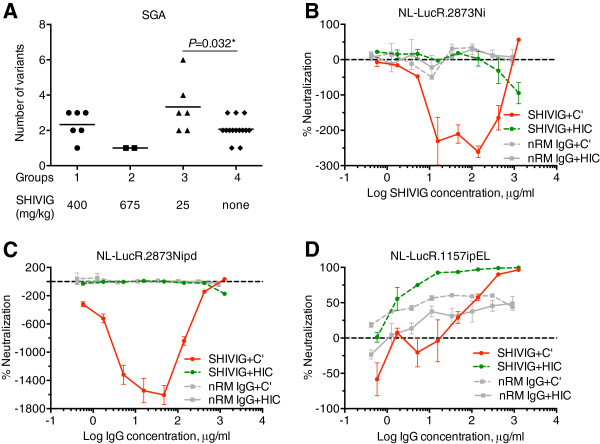
Figure 5.
Single-genome analysis (SGA) and C’-ADE. A. Number of SHIV-2873Nip quasispecies at peak viremia. The number of quasispecies was analyzed by SGA of the V1/V2 loop region of SHIV-2873Nip env. For every RM, at least 10 individual clones were obtained by limiting dilution PCR followed by sequencing. Sequence readout was performed using both strands of DNA. Statistical significance was assessed by Mann–Whitney test with Holm correction (P = 0.032). B-D. C’-ADE activity of SHIVIG in SupT1.R5 cells tested against different viruses built from the HIVNL4-3 backbone and containing the envelope of various SHIV or HIV-C strains as well as a Renilla luciferase (LucR) reporter gene. B. NL-LucR.2873Ni; C. NL-LucR.2873Nipd; and D. NL-LucR.1157ipEL. SHIVIG (red solid and green dashed lines) or nRM IgG (grey solid and dashed lines) were assayed in presence of 10% fresh (solid lines) or heat-inactivated (dashed lines) normal human serum as a source of complement. Each data point represents the mean ± SEM (n = 3). All experiments were repeated at least twice.