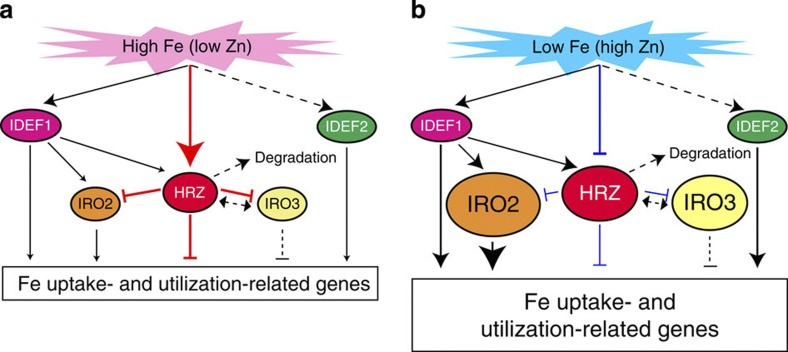
Figure 9. Hypothetical model of the signal transmission and regulation of Fe deficiency-inducible genes.
(a) Fe-sufficient conditions. (b) Fe-deficient conditions. In rice, the transcription factors IDEF1, IDEF2, OsIRO2 and OsIRO3 are known regulators of various Fe deficiency-inducible genes involved in Fe uptake and utilization5. IDEF1 binds to Fe and other metals (mainly Zn), and is thus proposed to be an Fe sensor that detects the cellular concentration ratio between Fe and other metals7. In the present study, we demonstrated that OsHRZ1 and OsHRZ2 also bind to Fe and Zn, presumably also serving as Fe sensors. HRZ proteins are likely to be rapidly degraded in roots irrespective of Fe status. HRZs negatively regulate the expression of various Fe deficiency-inducible genes involved in Fe uptake and utilization, mainly under conditions of Fe sufficiency (a). HRZs might function as modulators of the possible overexpression of Fe deficiency-inducible genes. On the other hand, HRZs may reduce or change their repression activity under Fe-deficient conditions as a consequence of the changes in Fe and Zn binding to haemerythrin domains (b). HRZs may indirectly interact with OsIRO3 because Arabidopsis BTS indirectly interacts with the basic helix–loop–helix transcription factor POPEYE8, which is a homologue of OsIRO3 (ref. 16), through the other basic helix–loop–helix transcription factors ILR3 and AtbHLH115. With the exception of IDEF1 and IDEF2, all of the indicated genes are transcriptionally induced under conditions of Fe deficiency (Fig. 2)5. The expression of HRZ genes is also positively regulated by IDEF1 (Supplementary Fig. S2). Thus, HRZs appear to form a negative feedback loop of Fe-deficiency signalling transmitted via IDEF1. However, gene expression analysis in HRZ-knockdown lines indicated that HRZs regulate both IDEF1-dependent and -independent genes (Table 1)26. Therefore, HRZs may also be involved in Fe sensing bypassing the IDEF1 pathway. The thickness of the lines and size of the arrowheads indicate the relative strength of the regulation. Putative or unverified pathways are indicated by broken lines.