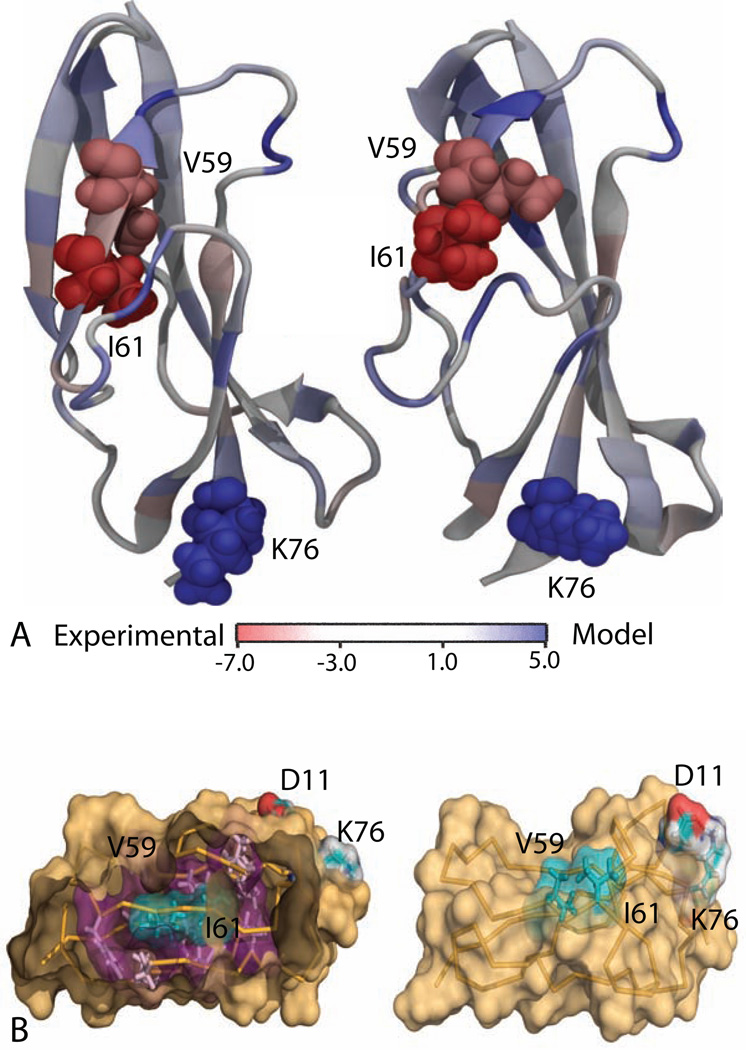
Figure 5.
(A) Per-residue free energy between the experimental NMR structure and the best prediction for CASP target T0569. The amino acid residues that are colored in deep red and deep blue stabilizes the NMR structure and the prediction, respectively; the residues with light blue color do not have a strong preference. (B) Key differences between the two structures as predicted by PRFE. The side chains of hydrophobic residues V59 and I61 are well packed and oriented towards the hydrophobic core in in the experimental structure (left) but they are exposed to the solvent in computer-generated model (right). A salt bridge between K76 and D11 stabilizes the computer-generated structure.