Figure 5.
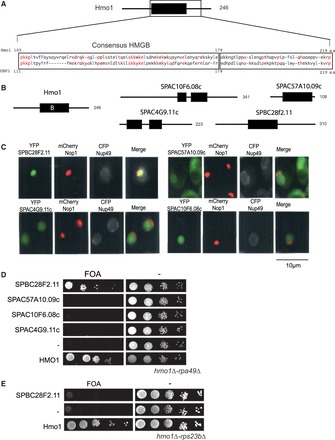
Identification of a Hmo1 counterpart in S. pombe. (A) Schematic representation of Hmo1. The position of the consensus HMGB region B (black rectangle) is shown in the schematic representation of Hmo1 and the amino-acid compositions of UBF1 and Hmo1 are presented in the close-up view. Residues that are identical are written in red and conserved hydrophobic residues are written in blue. (B) Localization of the HMGB of Hmo1 (black rectangle) and in four S. pombe’s proteins (Spbc28f2.11, Spac10f6.08c, Spac4g9.11c, Spac57a10.09c). Number of amino acids of each protein is indicated. (C) Spbc28f2.11 localizes to the nucleolus when expressed in budding yeast. The four HMGB proteins from S. pombe were co-expressed in S. cerevisiae as YFP fusion proteins (green) with CFP-Nup49 (gray) and mRFP-Nop1 (red) fusion proteins to visualize the nuclear periphery and the nucleolus, respectively (D) Spbc28f2.11 can substitute for the essential function of Hmo1 in a rpa49Δ-hmo1Δ background. Ten-fold serial dilutions of cultures of the rpa49Δ-hmo1Δ mutant expressing Hmo1, one HMGB from S. pombe or an empty vector (−) were spotted on plates containing 5-FOA to test for complementation. (E) Spbc28f2.11 cannot substitute for the essential function of Hmo1 in a hmo1Δ-rps23bΔ background. Ten-fold serial dilution of cultures of the rps23bΔ-hmo1Δ mutant expressing Hmo1, Spbc28f2.11 or an empty vector (−) were spotted on plates containing 5-FOA to test for complementation.
