Figure 6.
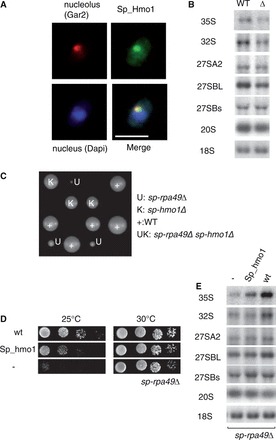
Characterization of the Hmo1 counterpart in S. pombe. (A) Localization of Sp-Hmo1 in S. pombe. Sp-Hmo1 was produced as a C-terminal HA-tagged protein in S. pombe and detected by immunofluorescence (green). The nucleolus was revealed by immunodetection of the Gar2 protein (red) and DAPI labeled the nucleus (blue). Scale bar 5 µm (B) Sp-Hmo1 is required for 35S pre-rRNA accumulation. Total RNA was extracted from a WT strain and Sp-hmo1Δ (Δ) mutant cells and analyzed by northern blotting. Hybridizations revealed the precursors indicated on the left of each panel. (C) The Sp-hmo1Δ-Sp-rpa49Δ double mutant is not viable. After mating of haploid Sp-hmo1Δ and Sp-rpa49Δ strains and meiosis, growth of segregants bearing Sp-hmo1Δ (K) and Sp-rpa49Δ (U) was followed by tetrad analysis. WT and single mutants Sp-hmo1Δ and Sp-Rpa49Δ were selected with KAN (K) and URA3 (U); their frequencies are close to those expected, but no double mutant was isolated. (D) Overexpression of Sp-Hmo1 suppresses the growth defect of Sp-rpa49Δ at 25°C. The Sp-rpa49Δ mutant transformed with empty vector (−), Sp-RPA49 complementing the deletion (WT) or Sp-HMO1 driven from a strong promoter were grown for 4 days at 25°C or 30°C. (E) Accumulation of rRNA precursors was restored in the Sp-rpa49Δ mutant by overexpression of Sp-Hmo1. Total RNA was extracted and analyzed by northern blotting. Hybridizations revealed the precursors indicated on the left of each panel.
