Figure 3.
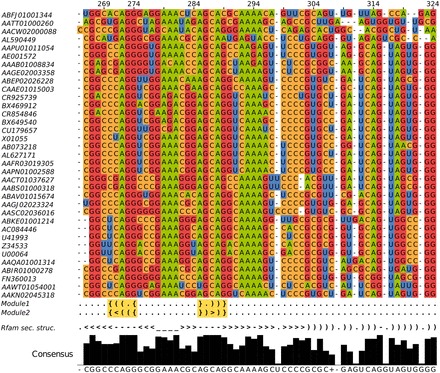
Rfam alignment with mapped modules. Two modules (Module 1 and Module 2) with different structures map on Rfam SRP family (RF00017cm) at position 270–274/285–289. Highlighted regions below the alignment show the module base pairs.’ ’ are single base pairs of canonical or non-canonical pairing type, whereas ‘
’ are single base pairs of canonical or non-canonical pairing type, whereas ‘ ’ denote bases pairing twice, i.e. to two nucleotides of type ‘
’ denote bases pairing twice, i.e. to two nucleotides of type ‘ ’ or ‘
’ or ‘ ’ [see (34)]. Rfam sec. struc. is the consensus secondary structure of Rfam. ‘
’ [see (34)]. Rfam sec. struc. is the consensus secondary structure of Rfam. ‘ ’ are used for ‘internal’ helices enclosing a multi-furcation of all terminal stems, ‘
’ are used for ‘internal’ helices enclosing a multi-furcation of all terminal stems, ‘ ’ show simple terminal stems, ‘
’ show simple terminal stems, ‘ ’ and ‘
’ and ‘ ’ denote unpaired bases of a hairpin loop and a bulge loop, respectively, and ‘.’ are insertions relative to a known structure. The histogram below shows the level of conservation of the alignment bases. The modules fit very well in the consensus secondary structure. Covariation information for model training can be obtained for four columns (270, 271, 285 and 290). The other columns are highly conserved. The consensus barplot below means the higher a bar the more conserved.
’ denote unpaired bases of a hairpin loop and a bulge loop, respectively, and ‘.’ are insertions relative to a known structure. The histogram below shows the level of conservation of the alignment bases. The modules fit very well in the consensus secondary structure. Covariation information for model training can be obtained for four columns (270, 271, 285 and 290). The other columns are highly conserved. The consensus barplot below means the higher a bar the more conserved.
