Figure 5.
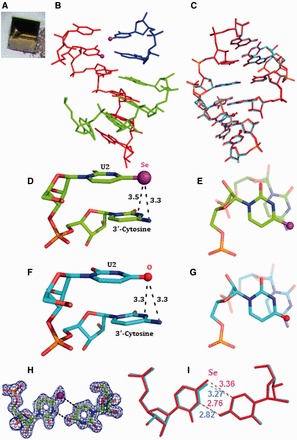
The yellow crystal and structures of the 4-Se-U RNA hexamer, (5′-U-SeU-CGCG-3′)2. The purple and red balls represent Se and O atoms, respectively. (A) The picture of the yellow Se-RNA crystal (0.1 × 0.1 × 0.1 mm). (B) Structure of the Se-RNA duplex containing the Se-RNA hexamer (in red), the base-paired CGCG (in green) and the U•U-paired U-SeU (in blue). (C) Superimposition of the Se-modified structure (in red; PDB ID: 3HGA; 1.30 Å resolution) and the native structure (in cyan; PDB ID: 1OSU; 1.40 Å resolution), the rmsd value is 0.09 Å. (D) Se-modified U2 stacks on its 3′-cytosine; the distance between the Se atom and exo-N4 of 3′-cytosine is 3.3 Å; the distance between the Se atom and C4 of 3′-cytosine is 3.5 Å. (E) is the top view of (D). (F) Native U2 stacks on its 3′-cytosine; the distance between the O atom and exo-N4 of 3′-cytosine is 3.3 Å; the distance between the O atom and C4 of 3′-cytosine is 3.3 Å. (G) is the top view of (F). (H) Electron density map (2Fo-Fc) and model of the SeU•U pair at the level of 1.0 σ. (I) Superimposition of SeU•U pair (in red) with native U•U pair (in cyan); the H-bond lengths are indicated individually.
