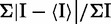Table 3.
Diffraction data collection and refinement statistics of the Se-RNA structures
| Structure (PDB ID) | U-SeU-CGCG (3HGA) | GUG-SeU-ACAC (4IQS) |
|---|---|---|
| Data collection | Se-Hexamer | Se-Octamer |
| Space group | C2221 | R32 |
| Cell dimensions: a,b,c (Å) | 30.255, 34.079, 28.931 | 47.006, 47.006, 354.105 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution range, Å (last shell) | 50.00–1.30 (1.32–1.30) | 50.0–2.60 (2.69–2.60) |
| Unique reflections | 3773 (162) | 8915 (846) |
| Completeness% | 95.9 (90.0) | 99.2 (95.8) |
| Rmerge% | 4.5 (26.1) | 5.3 (35.8) |
| I/σ(I) | 40.5 (1.2) | 35.9 (1.0) |
| Redundancy | 11.7 (4.2) | 10.0 (6.1) |
| Refinement | ||
| Resolution range, Å | 22.62–1.30 | 31.73.0–2.60 |
| Rwork% | 18.9 | 19.4 |
| Rfree% | 22.5 | 25.8 |
| Number of reflections | 3586 | 4776 |
| Number of atoms | ||
| Nucleic acid (single) | 157 | 1002 |
| Heavy atoms and ion | 1 Se | 6 Se |
| Water | 42 | 0 |
| R.m.s. deviations | ||
| Bond length, Å | 0.005 | 0.008 |
| Bond angle, ° | 0.931 | 1.846 |
Rmerge =