Figure 1.
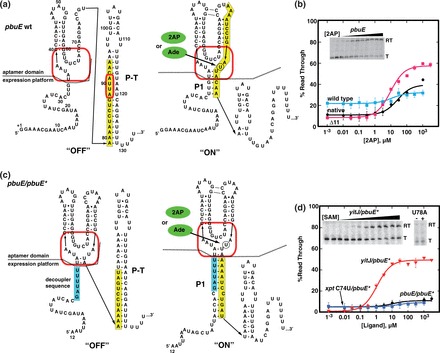
Reengineering the B. subtilis pbuE adenine riboswitch. (a) Sequence and secondary structure of the wild-type pbuE riboswitch in the ‘OFF’ and ‘ON’ states. The red box denotes the nucleotides comprising the ligand-binding pocket, and the yellow box denotes sequence predicted to adopt adenine-dependent alternative secondary structures. Numbering is consistent with the experimental determination of the transcription start site of this riboswitch (48). (b) In vitro assay of effector-dependent transcriptional anti-termination using 2AP for the native B. subtilis pbuE riboswitch transcriptional unit (‘native’), the wild-type pbuE riboswitch under control of the T7A1 promoter (‘wild type’) and the pbuE(Δ1–11) mutant (‘Δ11’). A representative denaturing gel showing 32P-labeled transcription products of the Δ11 mutant as a function of 2AP concentration is shown in the inset; ‘RT’ denotes the anti-terminated read through transcription product, and ‘T’ denotes the terminated product. Data plotted are the average of three independent measurements and the variation represented by the error bars. (c) Sequence and secondary structure of the pbuE ‘decoupled’ riboswitch capable of accommodating different aptamers. The cyan box denotes the insertion sequence that prevents the secondary structural switch from invading the aptamer domain. (d) In vitro transcription assay of the decoupled pbuE/pbuE* riboswitch along with two other aptamer chimeras. The inset demonstrates that the yitJ/pbuE* riboswitch regulates a transcriptional variation of the RNA and that a mutation (U78A) in the yitJ aptamer prevents SAM binding by the aptamer.
