Figure 4.
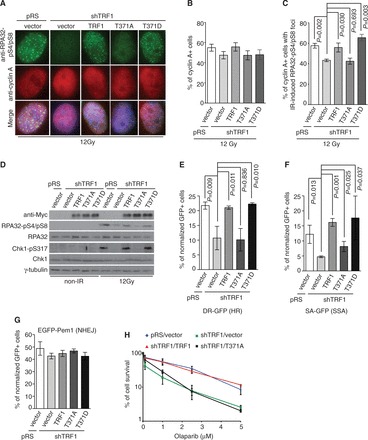
TRF1 phosphorylation at T371 facilitates DNA end resection and HR. (A) Indirect IF with anti-RPA32-pS4/pS8 in conjunction with anti-cyclin A antibody. TRF1-depleted HeLaII cells stably expressing the vector alone or various TRF1 alleles as indicated were treated with 12 Gy and fixed 8 h later. Cell nuclei were stained with DAPI in blue. (B) Quantification of cyclin A-positive cells in TRF1-depleted cells expressing various alleles as indicated. Standard deviations from six independent experiments are indicated. (C) Quantification of the percentage of cyclin A-positive cells with five or more IR-induced RPA32-pS4/pS8 foci from (A). Standard deviations from six independent experiments are indicated. (D) Western analysis of mock- or IR-treated TRF1-depleted HeLaII cells stably expressing the vector alone or various TRF1 alleles as indicated. Immunoblotting was carried out with anti-RPA32-pS4/pS8, anti-RPA32, anti-Chk1-pS317, anti-Chk1 or anti-γ-tubulin. (E) HR-mediated repair of I-SceI-induced DSBs. TRF1-depleted HeLaII cells expressing various TRF1 alleles as indicated were cotransfected with pDR-GFP, pCherry and I-SceI expression constructs. The number of cells positive for both GFP and pCherry was normalized to the total number of pCherry-positive cells, giving rise to the percentage of normalized GFP-positive cells. Standard deviations from four independent experiments are indicated. (F) SSA-mediated repair of I-SceI-induced DSBs. TRF1-depleted HeLaII cells expressing various TRF1 alleles as indicated were cotransfected with pSA-GFP, pCherry and I-SceI expression constructs. The quantification was done as described in (E). Standard deviations from three independent experiments are indicated. (G) NHEJ-mediated repair of I-SceI-induced DSBs. TRF1-depleted HeLaII cells expressing various TRF1 alleles as indicated were cotransfected with pEGFP-Pem1-Ad2, pCherry and I-SceI expression constructs. The quantification was done as described in (E). Standard deviations from three independent experiments are indicated. (H) Clonogenic survival assays of Olaparib-treated HeLaII cells stably expressing various alleles as indicated. Standard deviations from three independent experiments are indicated.
