Figure 2.
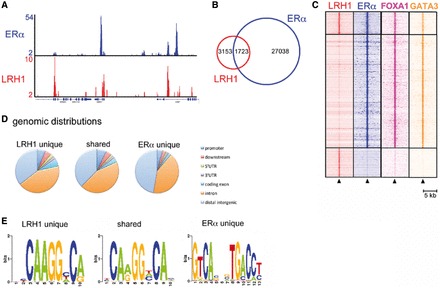
LRH-1 associates with ERα binding regions in chromatin. (A) ChIP-seq was carried out using HA and ERα antibodies for HA-LRH-1–transfected MCF-7 cells. Genome browser snapshot of ChIP-seq samples for ERα and HA-LRH-1 on proliferating cells. Y bar shows tag count. (B) Venn diagram of LRH-1 and ERα binding events. (C) Shown is a heatmap, with a window of 5 kb around the binding sites, depicting all shared and unique binding events for LRH-1 and ERα vertically aligned. Additionally, the binding events of the FOXA1 (24) and GATA3 (25) ChIP-sequencing data sets are shown. (D) The genomic distribution of shared and unique binding events. (E) De novo motif analysis of the shared and unique ERα and LRH-1 binding events.
