Figure 1.
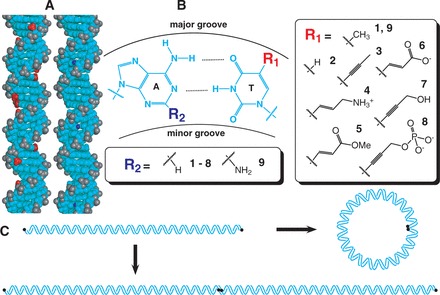
Experimental design. (A). Three-dimensional structure of B-form DNA (26). The central stacked DNA base pairs and deoxyribose sugars are uncharged (cyan), while each phosphodiester linkage carries a negative charge (gray). Seven of the eight tested base modifications replace the methyl group (red) of thymine bases in the DNA major groove, while one modification replaces the N2 proton (blue) of adenine bases in the DNA minor groove. (B). Chemical structure of an A·T base pair. Base modifications occur either at the 5 position of thymine (compare 1 with 2-8) or the 2 position of adenine (9). Based on the pKa values of the isolated functional groups, modifications 4, 6, and 8 alter DNA charge at neutral pH. (C). DNA ligase-catalyzed cyclization kinetics experiments to analyze DNA bend and twist stiffness. End-labeled (black circle) linear DNA fragments (∼200 bp, top) are detected when they either cyclize (right) or multimerize (bottom). The readout of these experiments is a ring closure probability (J-factor), which can be interpreted using the WLC model to estimate DNA mechanical parameters.
