Figure 4.
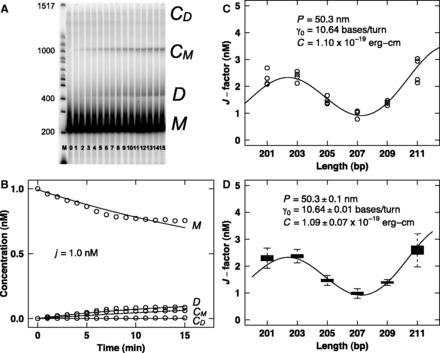
Example measurement of mechanical properties. (A) Cyclization time course for 207-bp DNA-like polymer 5 (pJ1744). DNA ligase-catalyzed cyclization reaction was performed at ∼22°C with 1 nM DNA restriction fragment, T4 DNA ligation buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 1 mM ATP, 10 mM dithiothreitol) and a final concentration of 100 U/ml T4 DNA ligase. Aliquots (10 µl) were removed at 1–15 min time points, quenched by addition of EDTA to 20 mM and then analyzed by electrophoresis through 5% native polyacrylamide gels in 0.5× TBE buffer (50 mM Tris base, 55 mM boric acid and 1 mM EDTA, pH 8.3). Gel lanes contains Invitrogen 100 bp DNA ladder (M), linear monomer without ligase (0) and increasing 1-min time points of the ligation reaction (1–15) showing the evolution of linear monomer (M), linear dimer (D), circular monomer (CM) and circular dimer (CD). Nearest molecular weight bands are indicated. (B) Cyclization kinetics analysis for 207-bp DNA-like polymer 5 (pJ1744). Fitting of data in (A) determines the J-factor, as previously described (30) (see also Supplementary Data S3). (C) WLC analysis for DNA-like polymer 5. Fit of experimental J-factor data using the WLC model. (D). Monte Carlo estimation of uncertainty. Fit of simulated J-factor data based on (C) using the WLC model.
