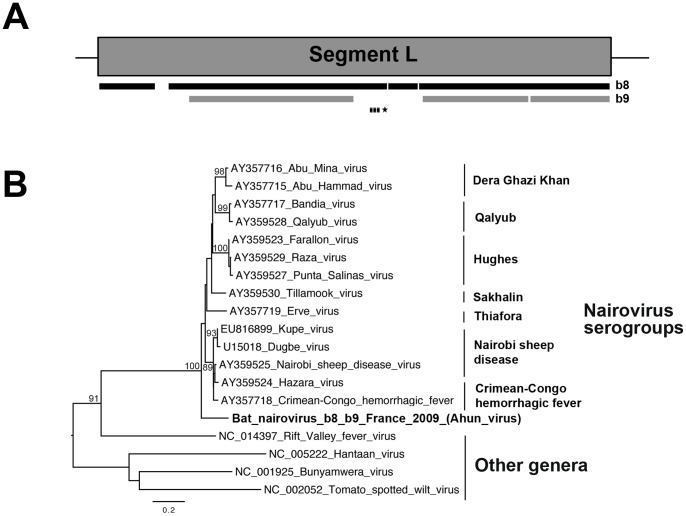
Figure 3. Phylogenetic analysis of the bat nairovirus-related sequences.
(A) Schematic representation of the large (L) segment (almost 12,260 nt encoding the RNA-dependent RNA polymerase of almost 4,040 aa) from the genome of Dugbe virus (GenBank number U15018), with black and gray bars corresponding to the longest contig sequences (>300 nt) of the bat nairovirus (named Ahun nairovirus) identified in specimens b8 and b9, respectively. The genomic region amplified by PCR is represented by a dashed bar, and the sequence used for phylogenetic analysis is indicated with an asterisk. (B) Phylogenetic tree produced from the amino-acid alignment of the partial polymerase fragment (396-nt sequence, translated into a 132-aa sequence, aa positions 2317 to 2448 of the L protein of Dugbe virus, and aligned in accordance with previous studies [86], [87]). The name of the bat nairovirus is indicated in bold. The various serogroups of nairoviruses and prototype species of the other genera belonging to the family Bunyaviridae are indicated on the right of the tree. The scale bar indicates branch length, and bootstrap values ≥70% are shown next to the relevant nodes. The tree is midpoint-rooted for purposes of clarity only.