Table 5. Estimated QTL effects from the full model for grain weight.
| Loci(i, j)a |
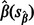 b
b
|
p-valuec |
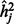 d
d
|
| (729_add, 729_add) | 1.02(0.07) | <10−15 | 0.1548 |
| (37_add, 547_dom) | 0.71(0.09) | 1.29×10−14 | 0.0428 |
| (67_add, 772_add) | −0.25(0.08) | 6.56×10−4 | 0.0047 |
| (96_add, 1117_dom) | 0.21(0.08) | 2.95×10−3 | 0.0035 |
| (119_add, 987_add) | 0.18(0.07) | 6.73×10−3 | 0.0024 |
| (151_add, 1262_add) | −0.15(0.07) | 9.79×10−3 | 0.0018 |
| (71_dom, 184_add) | −0.67(0.09) | 1.55×10−13 | 0.0374 |
| (210_add, 1400_add) | 0.19(0.08) | 7.79×10−3 | 0.0025 |
| (329_add, 727_dom) | 0.22(0.09) | 4.63×10−3 | 0.0040 |
| (310_dom, 419_add) | −0.21(0.05) | 1.86×10−5 | 0.0043 |
| (431_add, 1111_add) | 0.35(0.08) | 1.01×10−5 | 0.0107 |
| (71_dom, 500_add) | −0.76(0.08) | <10−15 | 0.0493 |
| (583_add, 1578_dom) | 0.35(0.08) | 9.50×10−6 | 0.0092 |
| (107_dom, 700_add) | 0.19(0.07) | 4.80×10−3 | 0.0035 |
| (708_dom, 714_add) | −1.15(0.32) | 1.79×10−4 | 0.0076 |
| (818_add, 1100_add) | 0.26(0.08) | 3.93×10−4 | 0.0053 |
| (916_add, 1026_add) | 0.15(0.06) | 8.50×10−3 | 0.0019 |
| (472_dom, 920_add) | −0.27(0.09) | 1.65×10−3 | 0.0058 |
| (18_dom, 955_add) | −0.20(0.08) | 3.77×10−3 | 0.0033 |
| (971_add, 1461_add) | 0.27(0.08) | 4.75×10−4 | 0.0075 |
| (620_dom, 1011_add) | −0.67(0.09) | 3.31×10−12 | 0.0336 |
| (1035_add, 1224_add) | 0.30(0.07) | 3.27×10−5 | 0.0081 |
| (1093_add, 1407_dom) | 0.44(0.08) | 2.13×10−7 | 0.0148 |
| (1167_dom, 1168_add) | −0.47(0.16) | 1.37×10−3 | 0.0051 |
| (119_dom, 1375_add) | 0.61(0.09) | 7.83×10−11 | 0.0289 |
| (1397_add, 1505_add) | 0.41(0.09) | 1.95×10−6 | 0.0119 |
| (247_dom, 1505_dom) | −0.23(0.08) | 1.22×10−3 | 0.0032 |
| (647_dom, 796_dom) | 0.26(0.08) | 4.29×10−4 | 0.0044 |
| Parameter(s) | a = 1, b = 1 | ||
| μ | −0.0661 | ||
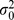
|
0.5317 | ||
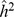
|
0.9379 | ||
aadd: additive effect; dom: dominance effect. If i equals j, then it is a main effect, otherwise, it is an interaction between locus i and locus j. Total number of effects is 90, only 28 effects with a p-value ≤0.01 are listed in this table.
bThe estimated marker effect is denoted by 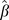 and the standard deviation is denoted by
and the standard deviation is denoted by 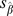 .
.
c P-value is obtained via t-test.
dPhenotypic variation explained.
