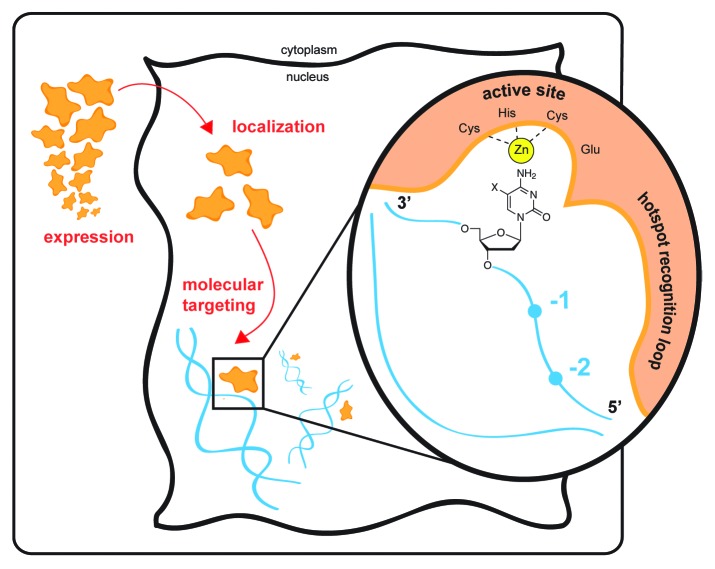
Figure 1. Levels of targeting of AID/APOBEC deaminases. Physiological targeting of the purposeful mutator AID/APOBEC enzymes occurs at many levels. Expression of AID is generally confined to the activated B cell, while APOBEC3 family restriction factors are largely expressed in myeloid and lymphoid lineages and, in certain settings, may be induced by inflammation or cytokine signaling. Beyond tissue-specific localization, cellular localization is critical for protecting genomic DNA from deaminases, and protein partners may be involved in engaging with genomic targets. At the molecular level, the enzymes preferentially engage with single-stranded DNA. Each family member shows a distinctive sequence preference that can span several residues surrounding the target base (−2 to 0 positions shown). The mutational signatures are conferred via the enzyme’s “hotspot recognition” loop that engages the target sequence. The active site, depicted with bound zinc (Zn) and the active site acid–base residue (Glu), engages with the cytosine base. The enzyme shows discrimination against 5-position modified cytosines (shown with an X). The deoxyribosyl sugar is a key determinant of proper targeting of DNA over RNA, facilitating a conformation which permits targeting of cytosine for deamination. These multiple levels of targeting govern the proper physiological engagement of substrate and perturbations at any of these levels may contribute to pathological pro-oncogenic mutation by AID/APOBEC enzymes.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.