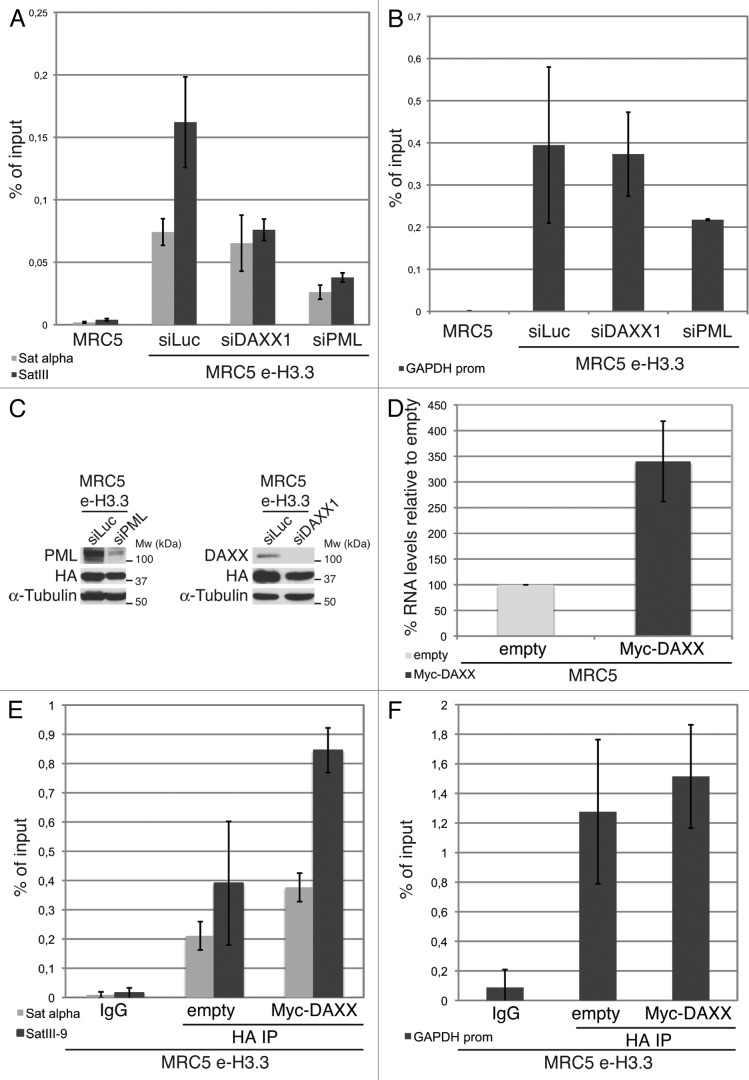
Figure 7.
Enrichment of H3.3 in pericentromeric heterochromatin is dependent on DAXX and PML proteins (A) Histogram shows analysis of H3.3 incorporation in centromeric (satellite α) or pericentromeric heterochromatin (satellite III) by ChIP against the HA epitope in MRC5 e-H3.3 cells treated for 72 h with the indicated siRNAs. QPCR data are presented as fold enrichment of IP over input DNA. MRC5 untransduced cells serve as a negative control. Numbers represent the mean of 2 independent experiments ± s.d. (B) Histogram shows analysis of H3.3 incorporation at the GAPDH promoter region by ChIP assay performed as in (A). Numbers represent the mean of 2 independent experiments ± s.d. (C) Western blot analysis of total cell extracts from MRC5 e-H3.3 cells treated as in (A). Membranes were probed for PML and DAXX to verify depletion of these proteins, and for HA to verify expression of the transduced proteins. α-tubulin is a loading control. M, molecular weight marker. (D) Satellite III mRNA expression levels in empty MRC5 cells or MRC5 cells overexpressing Myc-DAXX as determined by quantitative RT-PCR. mRNA levels were normalized to the reference gene GAPDH and levels were set to 100% in empty cells. Numbers represent the mean of 3 independent experiments ± s.d. (E) Histogram shows analysis of H3.3 incorporation in centromeric (satellite α) or pericentromeric heterochromatin (satellite III) by ChIP against the HA epitope in MRC5 e-H3.3 cells transduced with an empty vector (empty) or a vector encoding Myc-DAXX (Myc-DAXX). QPCR data are presented as fold enrichment of IP over input DNA. IP with IgG control antibody in MRC5 e-H3.3 cells serves as a negative control. Numbers represent the mean of 2 independent experiments ± s.d. (F) Histogram shows analysis of H3.3 incorporation at the GAPDH promoter region by ChIP assay performed as in (E). Numbers represent the mean of 2 independent experiments ± s.d.