Abstract
Two DNA vaccine plasmids encoding Herpes simplex virus type 2 (HSV-2) glycoprotein D, NTC8485-O2-gD2 and NTC8485-O2-UgD2tr, were produced at large scale under current good manufacturing practice (cGMP) for use in a Phase I human clinical trial. These DNA vaccines incorporate the regulatory agency compliant, minimal, antibiotic-free (AF) NTC8485 mammalian expression vector. Plasmid yields of > 1 g/L were achieved using the HyperGRO™ fed-batch fermentation process, with successful scale up from 10 L process development scale to 320 L culture volume for cGMP production. The DNA vaccines were purified using a low residence time, high shear lysis process and AIRMIXTM technology, followed by chromatographic purification. This combination of optimized plasmid vector, high yield upstream production, and efficient downstream purification resulted in purified HSV-2 DNA vaccines with > 99% total supercoiled plasmid, ≤ 0.2% RNA, ≤ 0.1% host cell genomic DNA, and ≤ 0.1 endotoxin units per mg.
Keywords: DNA vaccine, plasmid, Escherichia coli, antibiotic-free, HSV-2, fermentation, downstream processing
Introduction
As DNA vaccines progress in clinical trials, important factors should be considered for large scale cGMP manufacturing, including vector design, purity, concentration, and projected quantities needed upon approval and commercialization. Furthermore, to ensure safety, regulatory agencies recommend elimination of antibiotic resistance markers from therapeutic and vaccine plasmid DNA vectors.
Coridon Pty Ltd recently announced their Herpes Simplex Virus 2 (HSV-2) DNA vaccine to be 100% effective in protecting animals against infection (Allied Healthcare. 2010. Allied Healthcare announces Coridon license to vector technology [Press release]. Retrieved from http://www.asx.com.au/asxpdf/20120417/pdf/425n4kslf4lpyh.pdf). Herpes simplex virus type 2 (HSV-2) causes genital herpes and results in life-long infection. HSV-2 infections in newborns from infected mothers can have serious consequences,1 and HSV-2 might also aid the transmission of HIV.2 Treatment with antiviral drugs can suppress outbreaks, but does not prevent transmission of the disease. No approved HSV-2 vaccine currently exists.
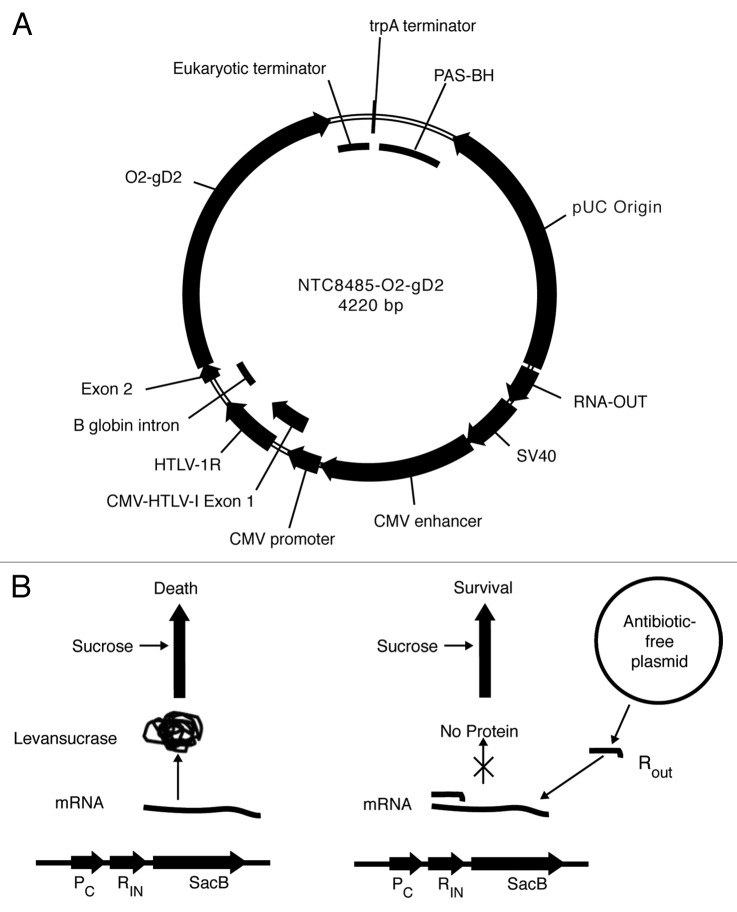
The Coridon DNA vaccines encode the envelope glycoprotein D sequence incorporated into the Nature Technology Corporation (NTC) NTC8485 antibiotic-free expression vector.3 This vector is specifically designed as a safe, minimalized, and antibiotic-free selection vector, which offers superior expression of recombinant proteins in mammalian cells, and is designed to comply with US Food and Drug Administration (FDA) and European Medicines Agency (EMA) regulatory guidance. The use of this vector improves the overall benefit of the vaccine by driving higher in vivo transcription and translation of the genetic material. Antibiotic-free selection in the NTC8485 vector is achieved by expression of a vector encoded 140 bp RNA-OUT antisense RNA that represses expression of a host strain chromosome-encoded counter-selectable marker (SacB) which is toxic when sucrose is present in the growth medium (Fig. 1B).
Figure 1. NTC antibiotic-free selection vectors. (A) NTC8485-O2-gD2 vector map; (B) Left: In a plasmid-free cell, levansucrase is expressed from a chromosomally integrated SacB gene, leading to cell death in the presence of sucrose. Right: RNA-OUT from the plasmid represses translation of the SacB gene, achieving plasmid selection.
In previous publications, NTC has reported results at the 10 L bioreactor scale using a patented, high cell density inducible fed-batch fermentation process (HyperGRO™) incorporating exponential nutrient feeding with a temperature shifting strategy, achieving plasmid yields up to 2.6 g/L culture in E. coli DH5α (Carnes and Williams, Process for plasmid DNA fermentation, 2011. US Patent 7 943 377).4,5 VGXI, Inc., a provider of DNA plasmid cGMP manufacturing and development services, has licensed NTC’s HyperGRO™ technology and successfully scaled-up cGMP fermentation using this process to manufacture Coridon’s HSV-2 DNA vaccines for a Phase I clinical trial. VGXI achieved reproducible plasmid yields of up to 1.8 g/L at the 320 L scale without the use of antibiotic selection, the largest fermentation scale yet using HyperGRO™ technology.
Following fermentation, plasmid DNA is typically isolated using an alkaline lysis-based process. The challenges of scaling up alkaline lysis are well known, most notably achieving sufficient mixing without damaging the plasmid DNA and creating difficult to remove host cell DNA fragments.6-8 VGXI has developed and patented large scale mixing processes for recovery of plasmid DNA by alkaline lysis, including a high-shear, low-residence time mixer for mixing the cell suspension with the alkaline lysis solution, and a bubble column mixer (AIRMIXTM) for neutralization of the lysate and separation of the flocculated host cell impurities (Hebel, et al. Devices and Methods for Biomaterial Production, 2007, US Patent 7 238 522).
Results and Discussion
Fermentation
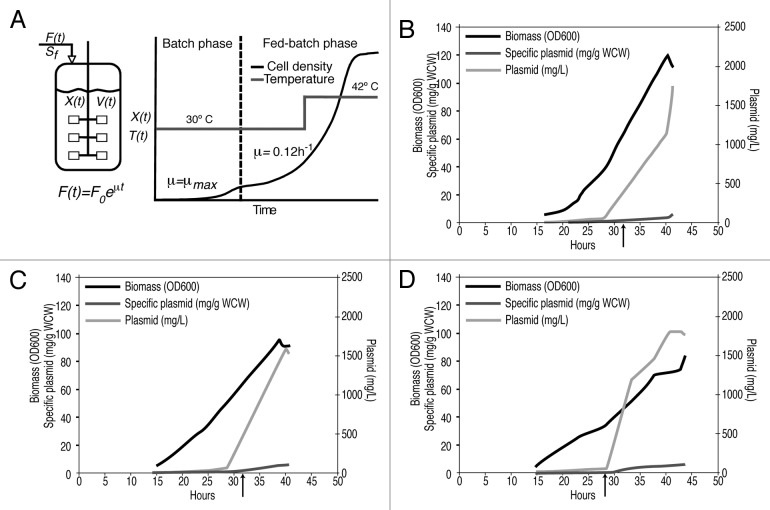
Very high yield plasmid production with the HyperGRO fed-batch process at the 10 L scale has been reported previously with temperature sensitive pUC origin type plasmids. These high yields result from nutrient limited fed-batch growth by an exponential feeding profile at 30 °C to accumulate approximately half of the total biomass, followed by a temperature shift to 42 °C for a period of several hours to induce high copy number plasmid amplification over the course of about one biomass doubling (Fig. 2A).
Figure 2. HyperGRO™ process: (A) process illustration; (B-D): Growth and plasmid yield profiles for HyperGRO™ fermentations with plasmid NTC8485-O2-gD2; the arrow marks the time of the temperature shift from 30 to 42 ºC: (B) 10L NTC process development fermentation; (C) 10L VGXI process development fermentation; (D) 320L VGXI cGMP fermentation.
For exponential feeding, the feed rate was controlled automatically by the bioreactor supervisory software using the following equation:9
F (t) = (μXBVB/SfYX/S) × eμt
where F(t) is the feed rate (L/h), μ is the desired specific growth rate during fed-batch phase (h−1), XB is the biomass concentration at the end of the batch phase (g dry cell weight/L), VB is the initial liquid volume of culture (L), Sf is the limiting substrate concentration in nutrient feed medium (g/L), YX/S is the yield coefficient of biomass from substrate (g/g) and t is the time since beginning of fed-batch phase (h). YX/S = 0.4 g/g, XB = 3.8 g/l and μ = 0.12 h−1 were used for the calculation of feed rate for all fed-batch fermentations. At both the 10 L process development scale and the 320 L cGMP scale, feeding was started 15 h following bioreactor inoculation.
Process development HyperGRO fermentations performed at the 10 L scale confirmed that the HSV-2 glycoprotein D antigen sequences had no deleterious effect on plasmid yield. Yields > 1 g/L were achieved, as expected from previous fermentations with the NTC8485 vector.10
High plasmid yields achieved at the 10 L process development scale were maintained at the 320 L cGMP scale (Table 1 and Fig. 2). Overall, the large scale cGMP fermentations resulted in pre-purification yields of approximately 586 g of plasmid NTC8485-O2-gD2 and 406 g of plasmid NTC8485-O2-UgD2tr.
Table 1. Yields of process development (10L) fermentations performed by NTC and VGXI and scaled up cGMP fermentations performed by VGXI.
| Fermentation scale | Plasmid | Final cell density (OD600) | Plasmid volumetric yield (mg/L) | Plasmid specific yield (mg/L/OD600) |
|---|---|---|---|---|
| NTC—10L (Fig. 2B) | NTC8485-O2-gD2 | 111 | 1700 | 15.3 |
| VGXI—10L (Fig. 2C) | NTC8485-O2-gD2 | 92 | 1530 | 16.7 |
| VGXI—320L (cGMP; Fig. 2D) | NTC8485-O2-gD2 | 85 | 1830 | 21.6 |
| VGXI—320L (cGMP) | NTC8485-O2-UgD2tr | 83 | 1270 | 15.4 |
Downstream purification
The downstream purification strategy was based on the final required amount of each component, not scaled to suit the entirety of fermentation biomass and specific yield.
Alkaline lysis using the proprietary AIRMIXTM technology followed by lysate clarification and subsequent binding to an anion exchange (AEX) membrane showed minimal loss through clarification steps and no detrimental effect on the plasmid product. In-process analysis showed that the bind and elute conditions allowed the flow through of impurities contained in the clarified lysate without compromising plasmid binding affinity. Due to the purification scale, the total binding capacity of the AEX membrane was exceeded at the approximate midpoint of the loading process. Consequentially, approximately 50% of the available plasmid DNA in the clarified lysate was bound to and recovered from the AEX membrane. The eluate from the AEX membrane contained 2400 mg (NTC8485-O2-gD2) and 2540 mg (NTC8485-O2-UgD2tr) of DNA with exceptionally low RNA content, not visible by agarose gel analysis.
Supercoil plasmid form enrichment was accomplished using hydrophobic interaction chromatography (HIC). The HIC load conditions allowed the retention of supercoiled plasmid DNA and certain contaminants while open circle plasmid forms and residual host cell DNA passed through the packed resin bed. The elute conditions resulted in the selective elution of supercoiled pDNA as evidenced by in process analysis and release testing of the plasmid product (Table 2). In both cases the dynamic binding capacity (2.5–2.8 mg/mL) was reached after approximately 50–60% of the conditioned load was processed, the remaining conditioned load was not passed through the column. 1090 mg (NTC8485-O2-gD2) and 1220 mg of (NTC8485-O2-UgD2tr) of pDNA were recovered in the HIC eluate.
Table 2. VGXI QC testing of purified plasmids NTC8485-O2-gD2 and NTC8485-O2-UgD2tr produced in scaled up cGMP HyperGRO fermentation at VGXI.
| Assay | Result | |
| NTC8485-O2-gD2 | NTC8485-O2-UgD2tr | |
| Nucleic acid concentration by A260, mg/mL | 2.8 | 2.8 |
| Purity by A260/280 | 1.9 | 1.9 |
| Identity by restriction analysis | CTS (conforms to standard) |
CTS (conforms to standard) |
| Structural integrity by percent circular forms analysis by HPLC, (total supercoil) % | 99 | 98 |
| Percent host-cell RNA, % | ≤ 0.2 | ≤ 0.2 |
| Percent host-cell protein, % | ≤ 0.1 | ≤ 0.1 |
| Percent host cell genomic, % | 0.1 | 0.004 |
| Endotoxin, EU/mg | ≤ 0.1 | ≤ 0.1 |
| Osmolality, mOsm/kgH2O | 37 | 27 |
| Visual appearance | Clear, colorless solution free from visible particulates | Clear, colorless solution free from visible particulates |
| pH | 7.7 | 7.7 |
| Plasmid identification - sequencing | No difference from reference sequence | No difference from reference sequence |
In order to maximize plasmid recovery from the UF/DF process a final nucleic acid concentration was targeted to over 150% of the final required concentration. Nucleic acid concentrations of 4.7 mg/mL (NTC8485-O2-gD2) and 6.2 mg/mL (NTC8485-O2-UgD2tr) were achieved with over 90% plasmid recovery.
Following dilution of the UF/DF product to the required concentration (2.8 mg/ml, Table 2) and aseptic processing, samples were analyzed by Quality Control. The plasmid product met all established limits for quality and purity assays; with up to 99% supercoiled plasmid and host cell impurities near the assay detection limit (Table2). The active bulk constituent from the purification processes, excluding filtered plasmid product for release testing, totaled 828 mg of NTC8485-O2-gD2 and 969 mg of NTC8485-O2-UgD2tr.
Using the plasmid mass and gel analysis from each step in purification an approximate yield of > 2.4 g of purified pDNA/ kg WCW can be inferred. If purification processes were scaled to suit the expected biomass and specific plasmid yield, a single 320 L HyperGRO fermentation followed by the VGXI purification process should yield a minimum of 175 g of each active bulk constituent for the HSV-2 DNA vaccine, which translates to approximately 40 000 filled units of HSV-2 vaccine.
Materials and Methods
Strains and plasmids
The plasmid host strain was E. coli NTC4862 (DH5α attλ::P5/6 6/6-RNA-IN- SacB, CmR).3 Plasmids NTC8485-O2-gD2 (Fig. 1A), 4220 bp, and NTC8485-O2-UgD2tr, 4190 bp were constructed using standard molecular biology methods. The NTC8485 vector contains a 140 bp DraIII-KpnI RNA-based sucrose selectable marker (RNA-OUT) and incorporates the high copy number PAS-BH-SV40 backbone from the kanR vector NTC7485.3,5
Fermentation
The HSV-2 DNA vaccine plasmids were transformed into E. coli NTC4862 individually, and prepared into research seed banks (RSB) and master cell banks (MCB). Fermentations were performed in semi-defined media according to the HyperGRO™ fed-batch fermentation process as described previously.4,5,11 Inocula were prepared from RSBs or MCBs and grown at 30 °C in seed medium containing 6% (w/v) sucrose. Batch phase fermentation media contained 5 g/L sucrose for plasmid selection. During fermentation, pH was controlled at 7.0 ± 0.1 by the automatic addition of 30% ammonium hydroxide or 10% phosphoric acid. The cultures were aerated at 0.5 to 1 VVM (volume of gas/volume of medium per min) and dissolved oxygen was maintained at 30% air saturation by proportional-integral control of agitation. Oxygen gas supplementation was initiated automatically as needed during fermentations to maintain dissolved oxygen at 30% air saturation. A semi-defined nutrient feed containing glycerol and yeast extract substrate was added according to a predetermined exponential feeding strategy to target a specific growth rate of µ = 0.12 h−1. Culture temperature was controlled at 30 °C until a cell density of 50 to 55 OD600 was reached, after which the temperature was shifted to 42 °C to induce high copy number plasmid amplification. Culture samples were taken at key points and at regular intervals during all fermentations. Samples were analyzed immediately for biomass (OD600) and for plasmid yield. Plasmid yield was determined by quantification of plasmid obtained from Qiagen Spin Miniprep Kit preparations as described.4 10L scale fermentations were performed at VGXI and NTC with a starting culture volume of 8L, in either New Brunswick Scientific BioFlo 3000 or BioFlo 110 bioreactors with a maximum working volume of 10.6 L. 320 L scale GMP fermentations were performed at VGXI with a starting culture volume of 220 L in a New Brunswick Scientific BioFlo Pro bioreactor with a 400 L maximum working volume.
Downstream purification process
Downstream processing of the HSV-2 DNA vaccine plasmids was performed by VGXI and consisted of cell lysis, solid/liquid separation, anion exchange membrane chromatography, hydrophobic interaction chromatography (HIC), ultrafiltration/diafiltration (UF/DF), and aseptic filtration. The cell lysis step was conducted using a proprietary lysis skid and AIRMIXTM technology (Hebel, et al. Devices and Methods for Biomaterial Production, 2007, US Patent 7 238 522) as previously described which was developed on the principle of alkaline lysis.8,12 The neutralized lysate was clarified with a decreasing pore size nylon monofilament mesh bag filter (Filter Specialist Inc.) followed by a 0.2 µm depth filter (Millipore) before being subjected to chromatography purifications. The anion exchange membrane chromatography employing Mustang Q capsules (Pall) captured the majority of plasmid DNA from crude lysate while allowing the majority of impurities to pass through. Plasmid product was eluted with a solution containing a higher ion concentration than the membrane equilibration. The eluate was conditioned with a high salt ammonium sulfate solution and loaded to an equilibrated XK 50/20 column (GE Healthcare) packed with Butyl 650M hydrophobic resin (Tosoh Biosep). Single step elution with a decreased concentration of ammonium sulfate was able to significantly reduce the open circular (OC) form of plasmid and selectively elute the SC plasmid, while separating residual contaminants. The recovered HIC product was further processed with an UF/DF step employing a disposable filter cassette (Novasep) of modified polyethersulfone membrane with a 50 kDa MWCO to displace the HIC elution buffer with 10 mM Tris (HCl) 1 mM EDTA pH 8 and increase the plasmid concentration to the target range. The UF product was filtered with 0.2 µm MiniKleenpak filters (Pall) and aliquoted for storage at -75 °C to -85 °C. In process analysis of purification samples was performed using nucleic acid quantification by A260 and 1% agarose gel analysis at all steps in the purification process.
Release testing of plasmid DNA product
The concentration of total nucleic acid in the product sample was measured with a SpectraMAX Plus 384 UV/Vis spectrophotometer (Molecular Devices) at the wavelength of 260nm, and the purity of plasmid DNA was evaluated with A260/A280. The osmolality of final plasmid was determined with the Advanced Micro-Osmometer Model 3300 (Advanced Instruments, Inc.). The host cell protein content was determined with a Micro BCA Protein Assay Reagent Kit (Pierce) using bovine serum albumin as the standard, and measuring the A562 with SpectraMAX PLUS 384 UV/Vis spectrophotometer (Molecular Devices). The endotoxin concentration in the final DNA product was measured with the Limulus Amebocyte Lysate (LAL) kinetic chromogenic assay (Charles River), and the A405 of samples was recorded with a SpectraMAX Pus 384 Microplate Reader. RNA quantification was performed by HPLC (Agilent 1100 Series) using a Butyl-NPR Column (Tosoh Biosep). The residual host cell DNA percentage was determined by Q-PCR using the Applied BioSystems Step One Plus Real Time PCR System). Plasmid isoform evaluation of final product was conducted by HPLC (Agilent 1100 Series), using a linear gradient with a TSKgel Butyl-NPR Column (Tosoh BioSep) to separate plasmid isoforms at A260.
Conclusion
VGXI’s successful collaboration with Nature Technology Corporation and Coridon Pty Ltd in the implementation and > 30 times scale up of the NTC HyperGRO™ antibiotic-free fermentation system has resulted in reproducible, high yield, plasmid producing fermentation under cGMP conditions for Coridon’s HSV-2 DNA vaccine. VGXI’s downstream purification process with plasmid bearing cells produced using the NTC HyperGRO™ process yielded plasmid product with supercoiled pDNA percentages of up to 99% and a very low contaminant profile (Table 2), providing high quality plasmid DNA for Coridon’s HSV-2 DNA vaccine Phase I clinical trial. In addition, it will provide a plasmid DNA manufacturing platform for future clinical trials which meets the strictest of regulatory demands.
Disclosure of Potential Conflicts of Interest
Jared Nelson and Stephen Rodriguez have a financial interest in VGXI, Inc. Neil Finlayson has a financial interest in Coridon Pty Ltd. Aaron E Carnes and James A Williams have a financial interest in Nature Technology Corporation.
Footnotes
Previously published online: www.landesbioscience.com/journals/vaccines/article/25048
References
- 1.Corey L, Wald A. Maternal and neonatal herpes simplex virus infections. N Engl J Med. 2009;361:1376–85. doi: 10.1056/NEJMra0807633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chentoufi AA, Dervillez X, Rubbo PA, Kuo T, Zhang X, Nagot N, et al. Current trends in negative immuno-synergy between two sexually transmitted infectious viruses: HIV-1 and HSV-1/2. Curr Trends Immunol. 2012;13:51–68. [PMC free article] [PubMed] [Google Scholar]
- 3.Luke JM, Vincent JM, Du SX, Gerdemann U, Leen AM, Whalen RG, et al. Improved antibiotic-free plasmid vector design by incorporation of transient expression enhancers. Gene Ther. 2011;18:334–43. doi: 10.1038/gt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carnes AE, Hodgson CP, Williams JA. Inducible Escherichia coli fermentation for increased plasmid DNA production. Biotechnol Appl Biochem. 2006;45:155–66. doi: 10.1042/BA20050223. [DOI] [PubMed] [Google Scholar]
- 5.Williams JA, Luke J, Langtry S, Anderson S, Hodgson CP, Carnes AE. Generic plasmid DNA production platform incorporating low metabolic burden seed-stock and fed-batch fermentation processes. Biotechnol Bioeng. 2009;103:1129–43. doi: 10.1002/bit.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoare M, Levy MS, Bracewell DG, Doig SD, Kong S, Titchener-Hooker N, et al. Bioprocess engineering issues that would be faced in producing a DNA vaccine at up to 100 m3 fermentation scale for an influenza pandemic. Biotechnol Prog. 2005;21:1577–92. doi: 10.1021/bp050190n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnes AE, Williams JA. Plasmid DNA manufacturing technology. Recent Pat Biotechnol. 2007;1:151–66. doi: 10.2174/187220807780809436. [DOI] [PubMed] [Google Scholar]
- 8.Hebel H, Attra H, Khan A, Draghia-Akli R. Successful parallel development and integration of a plasmid-based biologic, container/closure system and electrokinetic delivery device. Vaccine. 2006;24:4607–14. doi: 10.1016/j.vaccine.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 9.Lee SY. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 10.Carnes AE, Luke JM, Vincent JM, Anderson S, Schukar A, Hodgson CP, et al. Critical design criteria for minimal antibiotic-free plasmid vectors necessary to combine robust RNA Pol II and Pol III-mediated eukaryotic expression with high bacterial production yields. J Gene Med. 2010;12:818–31. doi: 10.1002/jgm.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carnes AE, Luke JM, Vincent JM, Schukar A, Anderson S, Hodgson CP, et al. Plasmid DNA fermentation strain and process-specific effects on vector yield, quality, and transgene expression. Biotechnol Bioeng. 2011;108:354–63. doi: 10.1002/bit.22936. [DOI] [PubMed] [Google Scholar]
- 12.Cai Y, Rodriguez S, Rameswaran R, Draghia-Akli R, Juba RJ, Jr., Hebel H. Production of pharmaceutical-grade plasmids at high concentration and high supercoiled percentage. Vaccine. 2010;28:2046–52. doi: 10.1016/j.vaccine.2009.10.057. [DOI] [PubMed] [Google Scholar]