Table 2.
Footprint sequences in DH and JH alleles.
| DH gene | Sequence (footprint(s) in red font) |
|---|---|
| D1-1*01 | GGTACAACTGGAACGAC |
| D1-14*01 | GGTATAACCGGAACCAC |
| D1-20*01 | GGTATAACTGGAACGAC |
| D1-26*01 | GGTATAGTGGGAGCTACTAC |
| D1-7*01 | GGTATAACTGGAACTAC |
| D2-15*01 | A TGTAGTGGTGGTAGCTGCTACTCC TGTAGTGGTGGTAGCTGCTACTCC |
| D2-2*01 | A TGTAGTAGTACCAGCTGCTATGCC TGTAGTAGTACCAGCTGCTATGCC |
| D2-2*02 | A TGTAGTAGTACCAGCTGCTATACC TGTAGTAGTACCAGCTGCTATACC |
| D2-2*03 | T TGTAGTAGTACCAGCTGCTATGCC TGTAGTAGTACCAGCTGCTATGCC |
| D2-21*01 | AGCATATTGTGGTGGTGATTGCTATTCC |
| D2-21*02 | AGCATATTGTGGTGGTGACTGCTATTCC |
| D2-8*01 | A TGTACTAATGGTGTATGCTATACC TGTACTAATGGTGTATGCTATACC |
| D2-8*02 | A TGTACTGGTGGTGTATGCTATACC TGTACTGGTGGTGTATGCTATACC |
| D3-10*01 | GTATTACTATGGTTCGGGGAGTTATTATAAC |
| D3-10*02 | GTATTACTATGTTCGGGGAGTTATTATAAC |
| D3-16*01 | GTATTATGATTACGTTTGGGGGAGTTATGCTTATACC |
| D3-16*02 | GTATTATGATTACGTTTGGGGGAGTTATCGTTATACC |
| D3-22*01 | GTATTACTATGATAGTAGTGGTTATTACTAC |
| D3-3*01 | GTATTACGATTTT TGGTTATTATACC TGGTTATTATACC |
| D3-3*02 | GTATTAGCATTTT TGGTTATTATACC TGGTTATTATACC |
| D3-9*01 | GTATTAC TTTGACTGGTTATTATAAC TTTGACTGGTTATTATAAC |
| D4-11*01 | TGACTACAGTAACTAC |
| D4-17*01 | TGAC TGACTAC TGACTAC |
| D4-23*01 | TGAC TGGTAACTCC TGGTAACTCC |
| D4-4*01 | TGACTACAGTAACTAC |
| D5-12*01 | GT GTGGCTACGATTAC GTGGCTACGATTAC |
| D5-18*01 | GT AGCTATGGTTAC AGCTATGGTTAC |
| D5-24*01 | G GGCTACAATTAC GGCTACAATTAC |
| D5-5*01 | GT AGCTATGGTTAC AGCTATGGTTAC |
| D6-13*01 | GGGTATAGCAGCAGCTGGTAC |
| D6-19*01 | GGGTATAGCAGTGGCTGGTAC |
| D6-25*01 | GGGTATAGCAGCGGCTAC |
| D6-6*01 | GAGTATAGCAGCTCGTCC |
| D7-27*01 | CTAACTGGGGA |
| JH gene | Sequence (footprint(s) in red font) |
| J1*01 | GCTGAATACTTCCAGCACTGGGGCCAGGGCACCCTGGTC ACCGTCTCCTCAG |
| J2*01 | CTACTGGTACTTCGATCTCTGGGGCCGTGGCACCCTGGTC ACTGTCTCCTCAG |
| J3*01 | TGATGCTTTTGATGTCTGGGGCCA CAATGGTCACCG TCTCTTCAG CAATGGTCACCG TCTCTTCAG |
| J3*02 | TGATGCTTTTGATATCTGGGGCCA CAATGGTCACCG TCTCTTCAG CAATGGTCACCG TCTCTTCAG |
| J4*01 | ACTACTTTGACTACTGGGGCCAAGGAACCCTGGTCACCGTCT CCTCAG |
| J4*02 | ACTACTTTGACTACTGGGGCC ACCCTGGTCACCGTCT CCTCAG ACCCTGGTCACCGTCT CCTCAG |
| J4*03 | GCTACTTTGACTACTGGGGCCA CCCTGGTCACCGTCT CCTCAG CCCTGGTCACCGTCT CCTCAG |
| J5*01 | ACAACTGGTTCGACTCCTGGGGCCAAGGAACCCTGGTCACC GTCTCCTCAG |
| J5*02 | ACAACTGGTTCGACCCCTGGGGCC ACCCTGGTCACCGTCTCCTCAG ACCCTGGTCACCGTCTCCTCAG |
| J6*01 | ATTACTACTACTAC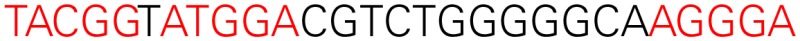 CCACGGTCACCGTCTCCTCAG CCACGGTCACCGTCTCCTCAG |
| J6*02 | ATTACTACTACTAC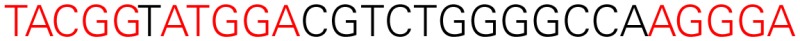 CCACGGTCACCGTCTCCTCAN CCACGGTCACCGTCTCCTCAN |
| J6*03 | ATTACTACTACTACTACTA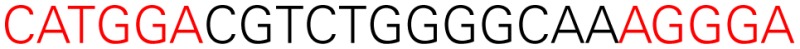 CCACGGTCACCGTCTCCTCAN CCACGGTCACCGTCTCCTCAN |
| J6*04 | ATTACTACTACTAC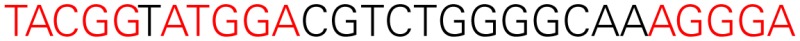 CCACGGTCACCGTCTCCTCAG CCACGGTCACCGTCTCCTCAG |
Thirty-four germline functional human DH alleles and 13 JH alleles were downloaded from the IMGT database (54). Each allele was scanned for all of the possible five nucleotide footprint motifs listed in Table 1. Sequences containing five nucleotide footprints are given in red font. In some cases the region matches more than one possible footprint (for example, D5-12*01 contains three different footprints: GGATA, GATAT, and ATATA).
