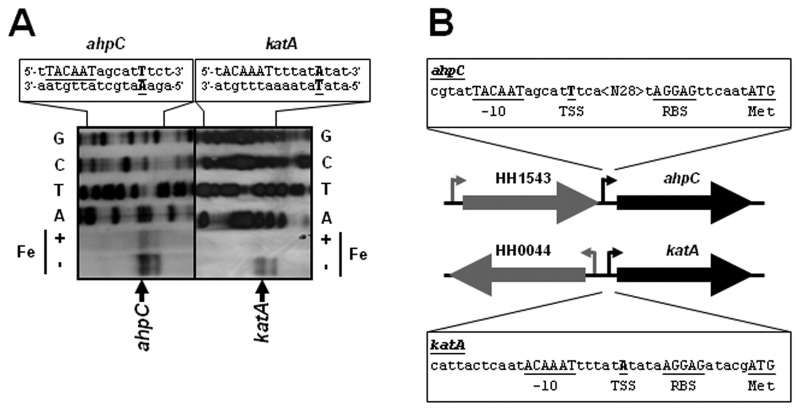
Fig. 1.
The H. hepaticus ahpC (HH1544) and katA (HH0043) genes have an iron-responsive promoter directly upstream of the coding sequence. (A) Primer extension analysis for the identification of the ahpC and katA transcription start sites. RNA from iron-restricted (–Fe) and iron-replete (+Fe) cultures of wild-type H. hepaticus ATCC 51449 was used for primer extension using a DIG-labeled primer (Table 1). Please note that the primer extension product displays iron-responsive repression of transcription. A standard dideoxy sequencing reaction was performed with the same DIG-labeled primer, and is shown above the primer extension reactions, with the respective dideoxynucleotide indicated next to the lanes, and the sequence of both strands given at the top of the figure. For both genes, the primer extension experiment resulted in three products, of which the product with the strongest band intensity is indicated by an arrow. For sequence annotation, see below. (B) Schematic representation of the genomic locations of the H. hepaticus ahpC and katA genes, with the transcription start site and associated promoter sequences annotated. The underlined sequence annotated as -10 is the sequence most closely resembling the σ70 –10 sequence (5’-TanaAT) identified in H. pylori [32]. The underlined residues represent annotated features in the sequence: TSS represents the transcription start site of the band highlighted in panel A, RBS is the proposed Shine-Dalgarno sequence, Met is the translational start of the gene, <N28> is a stretch of 28 nucleotides, and –10 is the proposed –10 box for the σ70 sigma factor