Abstract
Over the last 60 years, the use of hexachlorocyclohexane (HCH) as a pesticide has resulted in the production of >4 million tons of HCH waste, which has been dumped in open sinks across the globe. Here, the combination of the genomes of two genetic subspecies (Sphingobium japonicum UT26 and Sphingobium indicum B90A; isolated from two discrete geographical locations, Japan and India, respectively) capable of degrading HCH, with metagenomic data from an HCH dumpsite (∼450 mg HCH per g soil), enabled the reconstruction and validation of the last-common ancestor (LCA) genotype. Mapping the LCA genotype (3128 genes) to the subspecies genomes demonstrated that >20% of the genes in each subspecies were absent in the LCA. This includes two enzymes from the ‘upper' HCH degradation pathway, suggesting that the ancestor was unable to degrade HCH isomers, but descendants acquired lin genes by transposon-mediated lateral gene transfer. In addition, anthranilate and homogentisate degradation traits were found to be strain (selectively retained only by UT26) and environment (absent in the LCA and subspecies, but prevalent in the metagenome) specific, respectively. One draft secondary chromosome, two near complete plasmids and eight complete lin transposons were assembled from the metagenomic DNA. Collectively, these results reinforce the elastic nature of the genus Sphingobium, and describe the evolutionary acquisition mechanism of a xenobiotic degradation phenotype in response to environmental pollution. This also demonstrates for the first time the use of metagenomic data in ancestral genotype reconstruction, highlighting its potential to provide significant insight into the development of such phenotypes.
Keywords: hexachlorocyclohexane, metagenome, pan-genome, last-common ancestor
Introduction
Hexachlorocyclohexane (HCH) was one of the most popular pesticides for the control of agricultural pests and vector borne diseases during the latter half of the 20th century, and has subsequently become a key global pollutant culminating in the creation of open sinks with extremely high concentrations of HCH in the soil (for example, ∼450 mg g−1) (Jit et al., 2011).
HCH is prepared by the chlorination of benzene in the presence of UV, resulting in the formation of a technical HCH mixture, primarily containing five stable HCH isomers: α (60–70%), β (5–12%), γ (10–12%), δ (6–10%) and ɛ (3–4%) (Lal et al., 2010). Among these, only γ–HCH, also called lindane, has insecticidal properties and purification of one ton of γ–HCH results in 9–12 tons of HCH waste consisting primarily of α- and β-HCH. During the past six decades, ∼600 000 tons of γ-HCH had been generated; yielding 4–7 million tons of HCH waste (Lal et al., 2010). Apart from being carcinogenic, endocrine disruptors and neurotoxins (Kalantzi et al., 2004), these HCH isomers especially β and δ-HCH are highly persistent in the environment (Vega et al., 2007).
The first aerobic bacterium that could degrade lindane (γ-HCH), Sphingobium japonicum UT26, was isolated in 1989 from an HCH contaminated upland experimental field in Japan (Senoo and Wada, 1989). This was followed, in 1990, by the isolation of yet another sphingomonad, S. indicum B90A, from Indian rice rhizosphere soil repeatedly treated with technical HCH; this strain could degrade even the most recalcitrant HCH-isomer, β-HCH (Sahu et al., 1990). Over the past two decades both UT26 and B90A have served as models to explore the genetics and biochemistry of HCH-isomer degradation (Kumari et al., 2002; Dogra et al., 2004; Suar et al., 2004; Sharma et al., 2006; Nagata et al., 2011), and it is now well established that the degradation of HCH isomers in sphingomonads is mediated through enzymes encoded by lin genes (Lal et al., 2010). Among the lin genes, linA, linB and linC, which encode HCH dehydrochlorinase (LinA), haloalkane dehalogenase (LinB) and dehydrogenase (LinC), are responsible for the dehydrochlorination, hydroxylation and dehydrogenation of HCH isomers or their intermediates, respectively.
A recent metagenomic survey of a highly contaminated HCH dumpsite revealed (Supplementary Figures S1 and S2) the expected enrichment of Sphingomonadaceae, as well as lin genes, plasmids, and transposons over a gradient of increasing HCH contamination (Sangwan et al., 2012). To elucidate the ecology and evolution of the HCH-degrading phenotype in this ecosystem, we have reconstructed an in situ validated ancestral minimal genotype and provide evidence for the lateral gene transfer of xenobiotic degradative genes under the selective pressure of HCH pollution. This demonstrates for the first time the application of metagenomic data to ancestral genotype reconstruction, and highlights the potential of this approach to elucidate ecological genotype development, including demarcation between environmental and strain-specific traits.
Materials and methods
Soil and strain
Soil samples were collected from an open sink of HCH situated at Ummari village, Lucknow, India (27° 00′ 24.7′′ N, 81° 08′ 57.8′′ E) (Jit et al., 2011). As described earlier (Sangwan et al., 2012), subsamples (500 g soil per subsample) were collected at a depth range of 10–20 cm on 17 September 2011. Soil composition, sampling coordinates, methods and physicochemical analysis were performed as explained earlier (Sangwan et al., 2012). Laboratory strain of Sphingobium indicum B90A was used for the experiment.
DNA extraction and sequencing
Total DNA was isolated from each soil subsample (50 subsamples) using the PowerMax Soil DNA Isolation Kit (MO-BIO, Carlsbad, CA, USA). Equal concentrations (5 μg) of the environmental DNA from each subsample was mixed to form a homogenous composite genetic pool. DNA concentrations were quantified using NanoDrop spectrophotometer (NanoDrop Technologies Inc, Wilmington, DE, USA). Cell pellets from 5 ml pure culture of S. indicum B90A were subjected to genomic DNA isolation using QIAamp DNA Mini Kit (Qiagen, Venlo, Netherlands). DNA concentrations were quantified using NanoDrop spectrophotometer (NanoDrop Technologies Inc).
Paired-end reads were generated for genomic (3 μg μl−1) and metagenomic DNA (100 ng μl−1) using the 454 GS FLX Titanium (insert length=2000 bp±10, average read length: 320 bp±49) and the Illumina Genome Analyzer (San Diego, CA, USA) (insert length: 140 bp±6 and average read length, 75 bp±5) platforms, respectively. Illumina raw sequence data were subjected to various quality measures using the Seq-trim (Falgueras et al., 2010) pipeline and pyrosequencing output was analyzed using Roche 454 Analysis software version 2.0 (Branford, CT, USA). Reads below average score Q20, minimum length=50 bp -Illumina and 250 bp -pyrosequencing, and ambiguous bases (including N) were not included in any downstream analysis.
De-novo assembly of genome and metagenome
S. indicum B90A genome sequence reads (paired-end) were assembled into contigs using ABySS (Simpson et al., 2009) set at k-mer length of 41. Metagenomic reads (Table 1) were assembled by Metavelvet (Namiki et al., 2012) at k-mer length of 31, insert length=200 bp and expected coverage=auto. BAMBUS-2 (Koren et al., 2011) was used to generate metagenomic scaffolds. Assemblies (genome and metagenome) were validated and coverage was calculated by aligning the raw data (paired-end) back to the contigs using RTG Investigator (http://www.realtimemetagenomics.com). Any consensus mismatch between contig sequence and overlapping reads was considered as misassembly. Contigs from the previous metagenomic survey (Sangwan et al., 2012) were also used (minimum identity=95% and minimum overlap=50%) to bridge gaps between paired-end assembled metagenome contigs from this study.
Table 1. Sequencing and assembly statistics.
| Category | Sphingobium indicum B90A | Metagenome (Illumina data) |
|---|---|---|
| Sequence data | 450 Mbp (320 bp) | 1.6 Gbp (75 bp) |
| Reads after quality filtration | 42 044 290 | 20 111 630 |
| No. of contigs | 149 (>500 bp) | 1 216 300 (>200 bp) |
| Average contig coveragea | 80 | 3 |
| Reads used in assembly | 33 635 432 (80%) | 1 608 930(8%) |
| N50b | 95 Kbp | 253 bp |
| Max. contig size | ∼253 Kbp | ∼3 Kbp |
| GC content | 65 | 61 |
Abbreviation: N50 is the length of the smallest contig in a bin that contains largest contigs representing at least half of the total assembly length.
Calculated by mapping of reads to contigs with the criteria of 95% identity over 90% of the read length.
Based on minimum contig length criteria, that is, genome ⩾500 bp and metagenome ⩾200 bp.
Gene calling and annotations
Genes were predicted for S. indicum B90A draft genome using Glimmer-3 (Delcher et al., 2007). Metagenome contigs (minimum length=200 bp) were annotated at various hierarchy levels (individual enzymes, protein families, subsystem and cellular processes) using BLASTX (Altschul et al., 1990) (E-value=10−5) against COG (Tatusov et al., 2001), Pfam (Bateman et al., 2004) and KEGG (Kanehisa et al., 2004) databases. KEGG annotations were used in comparison against previous metagenomic predictions and hierarchical clustering was performed (n=1000) on the resultant matrix (sample versus category) using the pvclust package in R (R Development Core team, 2009). To taxonomically characterize and quantify the present genotypes in our metagenome data, individual metagenome reads were mapped over RefSeq (Pruitt et al., 2007) database (release 45, complete microbe) using RTG Investigator (http://www.realtimemetagenomics.com) (sequence identity cutoff=80%). Metagenomic 16S rRNAs were extracted (minimum length ∼150 bp) from contigs using BLASTN (Altschul et al., 1990) and compared against SILVA SSU rRNA database (Pruesse et al., 2007) using RTG investigator. SSU-align (Nawrocki and Eddy, 2010) was used to construct 16S rRNA domain (V3, V4 and V6) specific models corresponding to the bacterial 16S rRNA secondary structures and implementation of these domains in soil metagenome analysis (Vasileiadis et al., 2012). Metagenomic 16S rRNA sequences were aligned against these models and alignments were manually checked for any sequence repetition. Alignment files from RefSeq analysis were processed with SAMtools (Li et al., 2009) and total number of hits per reference sequence was calculated. Mapping results were further quantified into genera specific relative abundance using taxonomical information from their accession numbers. Pseudogenes were predicted in S. indicum B90A draft genome using Psi-Fi perl script (Guindon and Gascuel, 2003) and by manually checking (minimum identity mismatch=20%) the alignments for frame shift mutations. Pseudogene predictions were validated for sequencing errors by aligning the sequence reads back to the coding sequences (CDSs).
ANI and tetra nucleotide frequency calculations
Average nucleotide identity (ANI) values were calculated as explained in Konstantinidis and Tiedje (2005). All possible pairwise comparisons were performed between the S. indicum B90A draft genome (Anand et al., 2012) and the available Sphingobium genomes (chromosomes and plasmids). S. japonicum UT26 (Nagata et al., 2010), S. chlorophenolicum L-1 (Copley et al., 2012) and Sphingobium sp. SYK-6 (Masai et al., 2012) are the reference genomes of the organisms included in this study. Whole-genome-based tetranucleotide correlations were calculated using TETRA (Teeling et al., 2004). Euclidian distance matrix was constructed from ANI and tetranucleotide comparisons and hierarchical clustering was performed on the resultant matrix. These analyses were performed in R (R Development Core team, 2009) using ade (Dray and Dufour, 2007), pvclust and gclus packages.
Metagenomic recruitment of the genus Sphingobium
Metagenomic recruitment plots were generated for all available four genomes (chromosomes and plasmids) of the genus Sphingobium with MUMmer (Kurtz et al., 2004). Genomic coordinates covered (sequence identity and query coverage cutoff; 25% and 25% of the region, respectively) by metagenome reads tilling were selected as metagenomic islands (here after described as MGIs) (Steffen et al., 2012). Contigs from this study and previous dumpsite metagenome survey (Sangwan et al., 2012) were reassembled using Minimus (Sommer, 2007) at default parameter. We binned the metagenome contigs corresponding to reference genotypes (genus Sphingobium) using tetranucleotide frequencies (correlation cutoff (R2)=0.9) and %GC criterion (Tyson et al., 2004). BLASTN (Altschul et al., 1990) and MEGAN (Huson et al., 2007) were used to confirm the genetic identities of the contigs. ANI and tetranucleotide correlation values were also calculated for the ‘meta-Sphingobium' assembly (contigs) against complete Sphingobium reference genomes used in this study.
Identification and analysis of genomic and MGIs
Following the two step process to predict the complete genomic island profile, SIGI-HMM (Waack et al., 2006) algorithm (sensitivity value=0.7) was used to predict the potential genomic islands in ‘MGI free' (without MGIs) regions of the two genetic subspecies (ANI >98%). Genomic Islands were further annotated (BLASTX; E-value=10−5) against COG (Tatusov et al., 2001), KEGG (Kanehisa et al., 2004) and a local database created using protein CDSs from all Sphingobium genomes used in this study.
Reconstruction and comparative genomics of the ancestor genotype
To reconstruct the ancestor genotype of HCH-degrading subspecies, we used the complete genomes (chromosomes and plasmids) of S. indicum B90A and S. japonicum UT26 (ANI value of subspecies level ≥98% and both can degrade HCH isomers). As various Sphingobium spp. are known to carry the catabolic genes (lin genes) involved in HCH degradation, on their plasmids and secondary chromosomes (Dogra et al., 2004; Nagata et al., 2010; Tabata et al., 2011) we included the same of reference organisms for ancestor genotype estimation. Core genomic regions of S. indicum B90A and S. japonicum UT26 were separated from genomic islands using coordinates from metagenomic recruitment plots and SIGI-HMM predictions. Homologous anchors were computed in two subspecies using Murasaki (Popendorf et al., 2010) with seed weight-30 and seed length-40. Pre-computed homologous anchors were treated as input to predict orthologous segments using OSfinder algorithm (Hachiya et al., 2009). OSfinder predicts orthologous segments (syntenic) using Markov chain models and machine learning techniques. The advantage of using OSfinder lies in its automatic optimization of parameters to build markov models that increases the accuracy via minimizing the errors usually caused by setting the parameters manually (Pevzner and Tesler, 2003).
Orthologous regions (minimum ancestor genotype) were re-annotated with same strategy as explained above. Hypothetical proteins in the ancestor genotype were compared against the protein database of all available Sphingobium genomes. MEGABLAST (Zhang et al., 2000) was used to check the homologs of predicted foreign gene sequences (horizontally transferred) in ancestor genotype. Ancestral genotype (gene content) was mapped (BLASTN, E-value=10−10) against genome sequences of S. indicum B90A (Anand et al., 2012), S. japonicum UT26 (Nagata et al., 2010) and ‘meta-Sphingobium' assembly. Hierarchical clustering (minimum relative abundance=0.8% and s.d. cutoff=0.4%) was also performed on metagenome BLASTX results (Altschul et al., 1990) (E-value=10−5) analysis against KEGG (Kanehisa et al., 2004) database, using MeV 4.4 (Saeed et al., 2003) with Euclidean distance and Kendall's tau matrices and average linkage clustering.
Detection of genes under positive selection
Orthologous proteins from S. indicum B90A and S. japonicum UT26 were subsequently aligned, in a pairwise fashion using the CLUSTAL W algorithm (Thompson et al., 1994). The corresponding nucleotide sequences of these alignments were later aligned, codon by codon, using the pal2nal script (Suyama et al., 2006). Yn00 module of the PAML package (Yang, 2007) was used to calculate the dN/dS ratio for each pair of proteins. For proteins potentially involve in the degradation of phenol, toluene, chlorophenol, anthranilate, homogentisate and HCH, quality processed (quality score cutoff: Q15) dumpsite metagenome (pyrosequence data) reads were used to calculate dN/dS ratio using methodology as explained earlier (Tai et al., 2011).
Graph-based clustering and characterization of repetitive elements in genome and metagenome
Paired-end reads from this study, S. indicum B90A genome (insert size=2000±10 bp) dumpsite metagenome (insert size=140±6 bp) and pyrosequence reads from previous metagenome survey (Sangwan et al., 2012) were processed separately for de-novo identification and characterization of repetitive elements using RepeatExplorer pipeline (http://galaxy.umbr.cas.cz:8080). Briefly, assembly was performed using CAP3 (Huang and Madan, 1999) program with minimum overlap length for clustering=40% of the length (140 bp for genome and pyrosequence metagenome and 33 bp for Illumina metagenome) and minimum percentage identity criterion was set at 80. Results were analyzed using R program SeqGrapheR (Novak et al., 2010). Graph-based clustering (Novak et al., 2010) predicted 2223 (68% reads used) clusters in the whole genome and 817 870 (19% reads used) clusters in the metagenome. Clusters with similar annotations (minimum 40% percent overlap and 80% identity threshold) were merged and re-analyzed. Contigs (contributing in clusters) were annotated (BLASTX; E-value=10−5) against a custom database constructed using nucleotide sequences of insertion sequence (IS)-elements, integrases and putative tranposases present in the available genomes from genus Sphingobium and INTEGRALL database (Moura et al., 2009). To predict the genetic characteristics of transposon-mediated lateral transfer upon HCH contamination (insert size, expected sequences of insertion sites), we classified the predicted transposons to the ISfinder database (Siguier et al., 2006) using BLASTN (E-value=10−10, minimum identity 80% over 80% query length). Genetic fingerprints of the de-novo reconstructed transposons were traced back on the draft assembly of S. indicum B90A, the complete genome of S. japonicum UT26 and modern lineages using BLASTN (E-value=10−10).
Sequence availability
Draft assembly of S. indicum B90A is available in Genbank database with accession number AJXQ00000000. Metagenome sequence data have been deposited at DDBJ/EMBL/GenBank under the study accession number of ERP001726 (http://www.ebi.ac.uk/ena/data/view/ERP001726).
Results and discussion
Phylogenomics of the genus Sphingobium: species to ecotypes
A non-redundant database was created (14.98 Mb, 107 735 reads; clustering cutoff at pan-genome level 97% identity) containing HCH dumpsite metagenomic reads that mapped to four Sphingobium genomes. The relative abundance of HCH-degrading genotypes, S. indicum B90A and S. japonicum UT26, was collectively much higher (91 275 reads; 0.8% of the total metagenome data with ANI >98%) than the non-HCH-degrading genotypes of S. chlorophenolicum L-1 and Sphingobium sp. SYK-6 (16 460 reads; 0.3% of the total metagenome data with ANI >98%). The four Sphingobium species, demonstrated close phylogenetic similarity (16S rRNA nucleotide identity of 97–99.6%) (Pal et al., 2005), but to further establish the evolutionary kinship among these isolates, ANI values were computed with the secondary genetic elements, which often carry the lin genes (Nagata et al., 2010; Nagata et al., 2011; Tabata et al., 2011) (Supplementary Figure S3a). Phylogenomic sequence analysis of S. indicum B90A and S. japonicum UT26 revealed that they are genetic subspecies (ANI=98.04%); whereas Sphingobium sp. SYK-6 and S. chlorophenolicum L-1 are considerably less closely related (ANI=89%) as previously reported using 16S rRNA gene sequence similarity (Copley et al., 2012). Genetic relatedness was further validated using tetranucleotide profiling, which showed a similar relationship as defined by ANI analysis (Supplementary Figure S3b).
As the abundance of populations closely related to S. indicum B90A and S. japonicum UT26 (ANI >98%) was high (>0.8%) at the dumpsite, we hypothesized that their close relatives would have inherited a minimal ancestor gene complement. In order to validate this hypothesis, metagenomic sequence fragments were assembled, and the resulting contigs were characterized using tetranucleotide frequency correlation and %GC into Sphingobium related bins, which were then reassembled. This ‘meta-Sphingobium' assembly was, as expected, genetically (tetranucleotide frequencies and ANI) close to S. indicum B90A and S. japonicum UT26 (Supplementary Figure S3b). However, mapping the ‘meta-Sphingobium' assembly to the non-HCH-degrading Sphingobium strains (Sphingobium chlorophenolicum L1 and Sphingobium sp. SYK-6), clearly highlighted their genetic heterogeneity, corresponding to the higher relative abundance of their secondary genetic elements in comparison with the primary chromosome (Supplementary Figure S3a). The high relative pangenomic abundance (at species level identity cutoff of 97%) of plasmids and secondary chromosome genotypes within the dumpsite metagenome, suggests that both HCH-degrading and non-degrading Sphingobium strains have been selected for these secondary genetic elements in comparison with the gene content present on the primary chromosome, which highlights the selection pressure of the HCH contamination.
Recursive identification of ‘foreign genes'
The individual metagenomic reads (20 111 630 reads) from the dumpsite were mapped on to the draft genome of S. indicum B90A, as well as the two chromosomes (chromosome 1=3.5 Mb and chromosome 2=681.8 Kb) and three plasmids (pCHQ1=190.9 Kb, pUT1=31.7 Kb and pUT2=5.3 Kb) of S. japonicum UT26 (Supplementary Figure S3a). Regions with >90% sequence identity and a mapping coverage of ∼8 × were selected for ancestor genotype predictions (see next section), and those with low sequence identity (less than ∼25%) or low coverage (less than ∼25% of the region) were characterized as MGIs and further annotated to determine their strain or environment specific functional traits. By using this approach, we identified two MGI profiles; 631 genes (∼312 Kb) in the genome of S. indicum B90A and 822 genes (∼559 Kb) in the complete genome sequence of S. japonicum UT26.
Although, at a fine-scale (for example, KEGG enzyme) the functional profiles of the two MGIs were not very similar (R2=0.69 and P<0.0001; Fishers two-sided exact test with FDR correction; Supplementary Figure S3c), the majority of the genes still encoded for transposition, recombination and repair (B90A=15.094%±0.13 and UT=11.051%±0.01), as well as inorganic ions and amino-acid transport and metabolism (B90A=10.09%±0.18 and UT26=9.08%±1.4) (Supplementary Figure S3d). As, both the strains have been isolated from different locations (India and Japan), the differences in the functional potential of MGIs profiles could be attributed to the genetic adaptation of these strains in their microhabitats.
Besides the MGIs in both strains using the genomes were further analyzed for foreign genes using DNA composition characteristics via SIGI-HMM (Waack et al., 2006). The existing MGIs for each strain (predicted above) were validated using this algorithm. Although no further foreign genetic material was predicted for S. japonicum UT26 (Supplementary Table S2), the genome (excluding the MGI) of S. indicum B90A was predicted to contain 51 further foreign protein CDSs, totaling 31 Kb. These genes coded mostly for transposase, phage integrase, recombination and repair, and hypothetical proteins (Supplementary Figure S3d), the majority (89%±0.56) of which had greatest phylogenetic similarity to genes belonging to class Alphaproteobacteria. This demonstrates that metagenomic read mapping combined with DNA composition based algorithms (SIGI-HMM), provide a robust methodology for predicting a ‘complete' genomic island profile.
Reconstruction of the ancestral genotype and implications for legacy to recalcitrant compound degradation across modern lineages
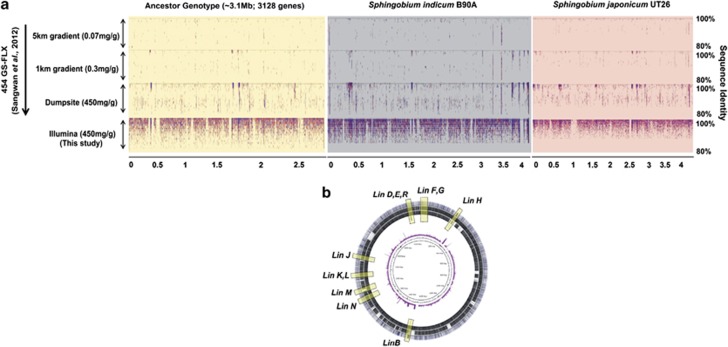
The minimal ancestral gene content inherited into each subspecies was reconstructed and its genetic fingerprint traced within the modern lineages. The metagenomic data (Table 1) was mapped (Supplementary Figure S3a) on to the genomes and plasmids of the two abundant subspecies (S. indicum B90A and S. japonicum UT26), and regions with >90% sequence identity and mapping coverage of >8 × were selected. The analysis was based on the assumption that a gene is ancestral if it was present in orthologous segments of the genetic subspecies (genomic regions without MGIs and SIGI-HMM predictions) and has abundant coverage in the metagenome. In all, a total of 3128 protein CDSs (3.04 Mb) were predicted for the minimal ancestral gene content inherited into sub-species from their immediate ancestor (Figures 1a and b). Putative CDSs from the predicted ancestral genotype were compared and mapped (BLASTN, E-value 10−8; Figure 1b) against the genomes of S. japonicum UT26 and S. indicum B90A and the available dumpsite metagenomic sequence data, to determine the relative genetic rearrangements across each discrete subspecies, and to trace the in situ relative abundance of these rearrangements in the metagenome (Figure 1b). Strikingly, although the lower HCH degradation pathway (linD, E, R, F, G, and H) was present in the ancestor, and abundant across all metagenomic data sets, there was a complete absence of two enzymes from the ‘core' upper HCH degradation pathway (linA, C genes) in the predicted ancestral genotype (Figure 1b). This suggests that the linA and linC were laterally acquired by both S. indicum B90A and S. japonicum UT26 (mean pairwise dN/dS >1, Supplementary Figure S4a).
Figure 1.
Predicted ancestral genotype, its metagenomic fingerprints and genetic rearrangements across S. indicum B90A and S. japonicum UT26. (a) Metagenomic fingerprints of ancestral gene content; Pyrosequence reads from the previous dumpsite metagenome survey (Sangwan et al., 2012) and illumina reads from this study were plotted on the predicted ancestral gene content (yellow shade) and genome sequences of the S. indicum B90A (gray shade) and S. japonicum UT26 (pink shade). Each dot on the graph represents an individual sequence read aligned along its reference genotypes. x and y axis represents the genomic coordinate (Mbp) and percentage sequence identity, respectively. (b) Genetic breakpoints between ancestor genotype and HCH-degrading genetic subspecies from outside towards centre; outermost circle: metagenomic contigs, circle 2: draft assembly of S. indicum B90A, circle 3: complete genome of S. japonicum UT26 mapped (BLASTN) over ancestral genotype (black color intensity represent the percentage identity, that is, darker the shade higher is the sequence identity), circle 4: lin genes homology between ancestor and subspecies, circle 5: metagenomic read coverage plot of ancestral gene content (5 × as coverage cutoff), outermost circle: GC plot of the predicted ancestral gene content.
Ecologically homogenous strains showing 1–5% genome-aggregate nucleotide divergence are already known to support the core genome hypothesis (Caro-Quintero and Konstantinidis, 2012) and sequence divergence within their genotypes corresponds to the evolution from the last-common ancestor (LCA). Thus, the fact that two major upper-pathway genes (linA and linC) of aerobic HCH degradation were not detected in the ancestral genotype, and that the HCH-degrading subspecies (Figure 1a and Supplementary Figure S4b) and the predicted ancestor demonstrate sequence-discrete population status (95–100% sequence similarity), suggests that the lower HCH degradation pathway (linD, linE and linR genes) is an evolutionary ‘long-lived' event (Caro-Quintero and Konstantinidis, 2012) in this ecosystem. This ‘long-lived' hypothesis is supported by the fact that the lower pathway genes were found in all metagenomic data sets (dumpsite, as well as 1 and 5 km from the dumpsite (Sangwan et al., 2012)), which is potentially explained by the close homology of linD, linE and linR to other catabolic genes also involved in biodegradation of other recalcitrant chemical compounds and present among several bacterial strains (Copley et al., 2012).
The estimated size of the ancestor genotype is 3 Mb (Figure 1a), which is smaller than that of the two subspecies (S. indicum B90A, 4.08 Mb and S. japonicum UT26, 4.4 Mb) and the estimated in situ bacterial specific average genome size (4.03 Mb±0.17) as predicted from the metagenomic data (Raes et al., 2007). The smaller size of an ancestral genotype has been reported to be a reliable indicator for large-scale lateral gene transfer in bacterial evolution (Dagan and Martin, 2007).
The core-metabolism of the ancestor (Supplementary Figure S4c) was similar to that of the sub-species (complete genomes), but strain-specific functional divergence (Supplementary Figure S4b) was also observed (ancestor versus S. sphingobium B90A, R2=0.99, P>0.0001; ancestor versus S. japonicum UT26, R2=0.78, P>0.0001). Raw reads from the genome of S. indicum B90A, the HCH dumpsite metagenome and the predicted ancestor's CDSs were all mapped on to the recalcitrant compound degradation pathway genes of S. japonicum UT26 (phenol/toluene, chlorophenol, anthranilate, homogentisate and HCH) (Nagata et al., 2011) (Supplementary Figure S4b). The data suggest that anthranilate degradation is a strain-specific character for S. japonicum UT26, inherited from an immediate ancestor, but completely absent in its genetic sub-species (S. indicum B90A) genome and in the in situ metagenome.
On the contrary, homogentisate degradation was found to be an environment specific trait (laterally acquired) as it was completely absent in the ancestor genotype and the S. indicum B90A (subspecies) genome (Supplementary Figure S5), but was present in the dumpsite metagenome. Sequence mapping analysis (identity >97%) (Supplementary Figure S4b), clearly revealed that phenol and chlorophenol degradation pathway genes were present in the ancestral genotype, the reference species, and their as-of-yet-uncultivated relatives present in the HCH dumpsite metagenome. This suggests the selective maintenance of this trait in the evolution of the sphingomonads exposed to HCH isomers.
As the ‘geographically distant' isolation sites of S. indicum B90A and S. japonicum UT26 did not share the physicochemical history of the analyzed site except amendment with HCH, these traits appear to be purely ancestral. However, these traits are under strong natural selection pressure (mean pairwise dN/dS >1) and the codon usage patterns are in agreement with the core usage of the reference strains. Following the ‘genome streamlining theory' (Giovannoni et al., 2005) genetic maintenance of these traits at high evolutionary cost still needs to be explored.
De-novo identification and characterization of mobile genetic elements in situ
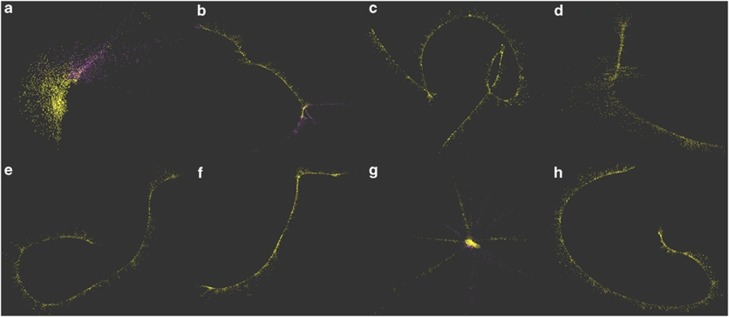
The absence of the upper HCH degradation pathway genes linA and linC in the ancestor generates the hypothesis that the sphingomonads were under strong selection pressure to acquire HCH degradation genes in HCH contaminated environments, and thereby increase their genome size. Therefore, both S. indicum B90A, S. japonicum UT26 and the dumpsite metagenome should have the appropriate number of mobile genetic elements, IS and integrases to support these predicted lateral gene transfer events (Aziz et al., 2010). To determine this, a de-novo approach was used to identify these elements in the genome of S. indicum B90A, and in the HCH dumpsite metagenome. A total of 19 (8.5%) clusters (Supplementary Figure S5a) from the genome and 267 (0.03%) from (Supplementary Figure S5b) the metagenome had significant similarities to the putative transposases and integrases. After merging overlapping clusters (contigs with minimum 40% percent overlap and 80% identity threshold), re-analysis and validation based on the metagenomic coverage cutoff (minimum coverage ∼8 × ), 8 unique clusters (representing complete transposases) were identified (Figure 2). All of the eight putative transposons were found to be from the IS6 family (IS6100).
Figure 2.
Metagenomic validation of de-novo assembled strain-specific mobile genetic elements. Total 8 transposons (a–h) were selected after reconstructed from de-novo assembly and graph-based clustering of genome (S. indicum B90A) and HCH dumpsite metagenomes. For each transposon; purple color dots represents the genomic reads of contigs (from graph-based clustering) assigned to a transposon and overlapping yellow reads represents the metagenomic validation with minimum sequence identity of 97%.
These results suggest that the eight transposons are preferentially environment specific, ecologically selected (1% to 5% pangenomic nucleotide divergence) and highly active (based on their abundance in the metagenomic data) mobile genetic elements and have been enriched in the bacterial lineages present at the dumpsite. Interestingly, on tracing these transposases back to the genomes of S. indicum B90A and S. japonicum UT26, four of them (Figures 2a) were found to be associated with linA, linA1, linC and linDER genes, respectively (Dogra et al., 2004; Nagata et al., 2010). These findings clearly explain the strong lateral transfer potential (Tn3 family) of lin genes in the reference (genomic) and as-yet-uncultured (metagenomic) HCH-degrading bacterial lineages. It also provides further mechanistic support for the hypothesis that linA and linC genes are environment specific genetic imports that were not present in the LCA of the two HCH-degrading subspecies.
Metagenomic recovery of enriched plasmids and a secondary chromosome
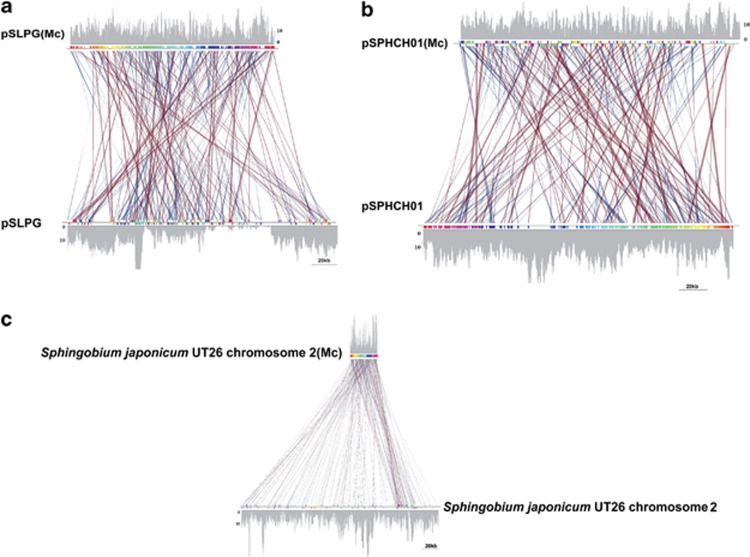
Whole-genome sequences of various Sphingobium species have revealed that most of the catabolic genes responsible for the degradation of xenobiotics are generally present on their plasmids (Nagata et al., 2011; Tabata et al., 2011; Copley et al., 2012). Comparative metagenomic analysis (BLASTN; E-value=10−8) against the NCBI plasmid database (ftp.ncbi.nlm.nih.gov/genomes/Plasmids/) revealed the enrichment of plasmids (P<0.001 Fisher's exact test with FDR correction) at the dumpsite (Supplementary Figure S5c), and increasing relative abundance across the increasing HCH contamination gradient (5 km <1 km <454 and Illumina dumpsite). For 37 enriched plasmid genotypes (Supplementary Figure S5c), a linear increase (P<0.0001) in metagenomic abundance was observed; 19 of which were more abundant in the Illumina data set compared with the 454-pyrosequenced metagenome (Supplementary Figure S5c), potentially as a result of the increased sequencing depth (Supplementary Table S1). Three draft genotypes were assembled from the metagenomic data (Figure 3 and Supplementary Figure S3a) corresponding to plasmids, pSPHCH01 (Copley et al., 2012), pSLGP (Masai et al., 2012), and chromosome 2 of S. japonicum UT26 (Lal et al., 2010; Dogra et al., 2004, Nagata et al., 2010). The relative enrichment of previously assembled plasmid variants (plasmid pLB1, pISP3 and pISP4; 12) were also validated. Nucleotide composition- (%GC and tetranucleotide frequency) based clustered contigs (species level structural variants) were arranged using paired end information and reference mapping (minimum percentage identity=85). Relatively similar nucleotide composition patterns were observed in pairwise comparisons between corresponding secondary genetic elements, their primary chromosomes and reconstructed genotypes (Copley et al., 2012).
Figure 3.
Genotypic synteny and metagenomic recruitment of reference genotypes. Whole-genome alignments between reference genomes and their reconstructed genotypes: (a) pSLPG plasmid of Sphingobium sp. SYK-6, (b) pSPHCH01 plasmid of Sphingobium chlorophenolicum L1 and (c) chromosome 2 of S. japonicum UT26. Overlaid the synteny plots are the metagenomic read coverage on a numerical scale. Mc, metagenome contigs.
Conclusions
The ‘genome streamlining theory' (Giovannoni et al., 2005), posits that selection pressure is linearly correlated with the effective genome size. Here, we have invoked this theory to explain the evolutionary increase in genome size as a result of selective pressure from xenobiotic pollution for microbial lineages present at an HCH dumpsite. Using nucleotide composition patterns, we have reanalysed the genetic relatedness among cultured representatives and in situ cohorts of the genus Sphingobium, and have shown that use of metagenomic data and combinatorial bioinformatic algorithms can provide a robust methodology for accurate ‘foreign gene' identification. Coherent genetic relatedness within ecotypes and their meta-pangenomic abundance has enabled us to predict the ancestral gene content inherited into their genotypes (strains). Furthermore, this study also proposes that metagenomics has the potential to clearly demarcate between environment and/or strain-specific functional traits acquired by closely related, sequenced reference organisms. These results suggest that we were successful in reconstructing an LCA genotype (as opposed to a core genome) as we were able to (a) clearly demark gene regions that were not part of the two reference genomes, (b) include secondary genetic elements and (c) identify and remove ‘forigen' genes using an LCA-specific algorithm. Strikingly, we found that despite the absence of various xenobiotic chemicals, for example, phenol, toluene, homogentisate and anthranilate, at HCH dumpsite, there is an evolutionary maintenance (from last ancestor to the cultured and yet-uncultivated cohorts) and positive selection of their biodegradation traits. The reasons for this are unclear, but it represents the powerful potential of metagenome-based ancestral genotype reconstruction for uncovering potentially important cross-linked genotype traits in such processes. Further genome sequencing of relevant isolates, and metagenomic sequencing and genome reconstruction from related sites, combined with knock-out mutants can now be targeted towards these processes and genes to help further elucidate the genetic mechanism.
Acknowledgments
The work was supported by Grants from the University of Delhi/Department of Science and Technology Promotion of University Research and Scientific Excellence (PURSE)-DU-DST—PURSE GRANT Department of Biotechnology (DBT), Government of India under project BT/PR3301/BCE/8/875/11, National Bureau of Agriculturally Important Microorganisms (NBAIM) AMASS/2006-07/NBAIM/CIR. NS, VN, HV and RK gratefully acknowledge Council for Scientific and Industrial Research (CSIR) and National Bureau of Agriculturally Important Microorganisms (NBAIM) for providing research fellowships. This work was also supported in part by the US Department of Energy under Contract DE-AC02-06CH11357.NS. We thank Konstantinos T Konstantinidis of School of Civil and Environmental Engineering and School of Biology, Georgia Institute of Technology, Atlanta, USA for his invaluable discussions and reading the manuscript. This paper was revised during a renewed visit under an Alexander von Humboldt Fellowship (at the University of Freiburg, Germany) awarded to RL.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anand S, Sangwan N, Lata P, Kaur J, Dua A, Singh AK, et al. Genome sequence of Sphingobium indicum B90A, a hexachlorocyclohexane-degrading bacterium. J Bacteriol. 2012;194:4471–4472. doi: 10.1128/JB.00901-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R-K, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucl Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, Konstantinidis KT. Bacterial species may exist, metagenomics reveal. Environ Microbiol. 2012;14:347–355. doi: 10.1111/j.1462-2920.2011.02668.x. [DOI] [PubMed] [Google Scholar]
- Copley SD, Rokicki J, Turner P, Daligault H, Nolan M, Land M. The whole genome sequence of sphingobium chlorophenolicum L-1: insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol Evol. 2012;4:184–198. doi: 10.1093/gbe/evr137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci USA. 2007;104:870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra C, Raina V, Pal R, Suar M, Lal S, Gartemann KH, et al. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J Bacteriol. 2004;186:2225–2235. doi: 10.1128/JB.186.8.2225-2235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour A. The ade4 package: implementing the duality diagram for ecologists. J Stat soft. 2007;22:1–20. [Google Scholar]
- Falgueras J, Lara AJ, Fernandez-Pozo N, Canton FR, Perez-Trabado, Claros MG, et al. SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinform. 2010;11:38. doi: 10.1186/1471-2105-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hachiya T, Osana Y, Popendorf K, Sakakibara Y. Accurate identification of orthologous segments among multiple genomes. Bioinformatics. 2009;25:853–860. doi: 10.1093/bioinformatics/btp070. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jit S, Dadhwal M, Kumari H, Jindal S, Kaur J, Lata P, et al. Evaluation of hexachlorocyclohexane contamination from the last lindane production plant operating in India. Environ Sci Pollut Res Int. 2011;18:586–597. doi: 10.1007/s11356-010-0401-4. [DOI] [PubMed] [Google Scholar]
- Kalantzi OI, Hewitt R, Ford KJ, Cooper L, Alcock RE, Thomas GO, et al. Low dose induction of micronuclei by lindane. Carcinogenesis. 2004;25:613–622. doi: 10.1093/carcin/bgh048. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Treangen TJ, Pop M. Bambus 2: scaffolding metagenomes. Bioinformatics. 2011;27:2964–2971. doi: 10.1093/bioinformatics/btr520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R, Subudhi S, Suar M, Dhingra G, Raina V, Dogra C, et al. Cloning and characterization of lin genes responsible for the degradation of hexachlorocyclohexane isomers by Sphingomonas paucimobilis strain B90. Appl Environ Microbiol. 2002;68:6021–6028. doi: 10.1128/AEM.68.12.6021-6028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal R, Pandey G, Sharma P, Kumari K, Malhotra S, Pandey R, et al. The biochemistry of microbial degradation of hexachlorocyclohexane (HCH) and prospects for bioremediation. Microbiol Mol Biol Rev. 2010;74:58–80. doi: 10.1128/MMBR.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E, Kamimura N, Kasai D, Oguchi A, Ankai A, Fuki S, et al. Complete genome sequence of Sphingobium sp. Strain SYK-6, a degrader of lignin-derived biaryls and monoaryls. J Bacteriol. 2012;194:534–535. doi: 10.1128/JB.06254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Natsui S, Endo R, Ohtsubo Y, Ichikawa N, Ankai A, et al. Genomic organization and genomic structural rearrangements of Sphingobium japonicum UT26, an archetypal γ-hexachlorocyclohexane-degrading bacterium. Enzyme Microb Technol. 2011;49:499–508. doi: 10.1016/j.enzmictec.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Ohtsubo Y, Endo R, Ichikawa N, Ankai A, Oquchi A, et al. Complete genome sequence of the representative γ-hexachlorocyclohexane-degrading bacterium Sphingobium japonicum UT26. J Bacteriol. 2010;192:5852–5853. doi: 10.1128/JB.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki T, Hachiya T, Tanaka H, Sakakibara Y. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012;40:e155. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.2010SSU-align: a tool for structural alignment of SSU rRNA sequencesAvailable at http://selab.janelia.org/software.html .
- Novak P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformat. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Bala S, Dadhwal M, Kumar M, Dhingra G, Prakash O, et al. Hexachlorocyclohexane-degrading bacterial strains Sphingomonas paucimobilis B90A, UT26 and Sp+, having similar lin genes, represent three distinct species, Sphingobium indicum sp. nov., Sphingobium japonicum sp. nov. and Sphingobium francense sp. nov., and reclassification of (Sphingomonas) chungbukensis as Sphingobium chungbukense comb. nov. Int J Syst Evol Microbiol. 2005;55:1965–1972. doi: 10.1099/ijs.0.63201-0. [DOI] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 2003;13:37–45. doi: 10.1101/gr.757503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popendorf K, Tsuyoshi H, Osana Y, Sakakibara Y. Murasaki: a fast, parallelizable algorithm to find anchors from multiple genomes. PLoS One. 2010;5:e12651. doi: 10.1371/journal.pone.0012651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sahu SK, Patnaik KK, Sharmila M, Sethunathan N. Degradation of alpha-, beta-, and gamma-hexachlorocyclohexane by a soil bacterium under aerobic conditions. Appl Environ Microbiol. 1990;56:3620–3622. doi: 10.1128/aem.56.11.3620-3622.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan N, Lata P, Dwivedi V, Singh A, Niharika N, Kaur J, et al. Comparative metagenomic analysis of soil microbial communities across three hexachlorocyclohexane contamination levels. PLoS One. 2012;7:e46219. doi: 10.1371/journal.pone.0046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo K, Wada H. Isolation and identification of an aerobic γ-HCH decomposing bacterium from soil. Soil Sci Plant Nutr. 1989;35:79–87. [Google Scholar]
- Sharma P, Raina V, Kumari R, Malhotra S, Dogra C, Kumari H, et al. Haloalkane dehalogenase LinB is responsible for β- and δ-hexachlorocyclohexane transformation in Sphingobium indicum B90A. Appl E Microbiol. 2006;72:5720–5727. doi: 10.1128/AEM.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I, et al. ABySS: a parallel assembler for short read sequence Data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer DD. Minimus: a fast, lightweight genome assembler. BMC Bioinformat. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen MM, Li Z, Effler TC, Hauser LJ, Boyer GL, Wilhelm SW. Comparative metagenomics of toxic freshwater cyanobacteria bloom communities on two continents. PLoS One. 2012;7:e44002. doi: 10.1371/journal.pone.0044002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suar M, van der Meer JR, Lawlor K, Holliger C, Lal R. Dynamics of multiple lin gene expression in Sphingomonas paucimobilis B90A in response to different hexachlorocyclohexane isomers. Appl Environ Microbio. 2004;70:5650–5656. doi: 10.1128/AEM.70.11.6650-6656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata M, Endo R, Ito M, Ohtsubo Y, Kumar A, Tsuda M, et al. The lin genes for γ—Hexachlorocyyclohexane degradation in Sphingomonas sp. MM-1 proved to be dispersed across multiple plasmids. Biosci Biotechnol Biochem. 2011;75:466–472. doi: 10.1271/bbb.100652. [DOI] [PubMed] [Google Scholar]
- Tai V, Poon AFY, Paulsen IT, Palenik B. Selection in coastal synechococcus (cyanobacteria) populations evaluated from environmental metagenomes. PLoS One. 2011;6:e24249. doi: 10.1371/journal.pone.0024249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtscev IV, Tautsov TA, Shankavaram UT, Rao BS, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Waldmann J, Lombardot T, Bauer M, Glöckner FO. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapmen J, Hugnholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Vasileiadis S, Puglisi E, Arena M, Cappa F, Cocconcelli PS, Trevisan M. Soil bacterial diversity screening using single 16S rRNA gene V regions coupled with multi-million read generating sequencing technologies. PLoS One. 2012;7:e42671. doi: 10.1371/journal.pone.0042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega FA, Covelo EF, Andrade ML. Accidental organochlorine pesticide contamination of soil in Porrino, Spain. J Environ Qual. 2007;36:272–279. doi: 10.2134/jeq2006.0053. [DOI] [PubMed] [Google Scholar]
- Waack S, Keller O, Asper R, Brodaq T, Damm C, Fricke WF, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformat. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.