Abstract
Arbuscular mycorrhizal fungi (AMF) are among the most abundant symbionts of plants, improving plant productivity and diversity. They are thought to mostly grow vegetatively, a trait assumed to limit adaptability. However, AMF can also harbor genetically different nuclei (nucleotypes). It has been shown that one AMF can produce genotypically novel offspring with proportions of different nucleotypes. We hypothesized that (1) AMF respond rapidly to a change of environment (plant host) through changes in the frequency of nucleotypes; (2) genotypically novel offspring exhibit different genetic responses to environmental change than the parent; and (3) genotypically novel offspring exhibit a wide range of phenotypic plasticity to a change of environment. We subjected AMF parents and offspring to a host shift. We observed rapid and large genotypic changes in all AMF lines that were not random. Genotypic and phenotypic responses were different among offspring and their parents. Even though growing vegetatively, AMF offspring display a broad range of genotypic and phenotypic changes in response to host shift. We conclude that AMF have the ability to rapidly produce variable progeny, increasing their probability to produce offspring with different fitness than their parents and, consequently, their potential adaptability to new environmental conditions. Such genotypic and phenotypic flexibility could be a fast alternative to sexual reproduction and is likely to be a key to the ecological success of AMF.
Keywords: arbuscular mycorrhizal fungi, genotypic plasticity, phenotypic plasticity, segregation, host shift, symbiosis
Introduction
Arbuscular mycorrhizal fungi (AMF) are among the most abundant symbionts worldwide. They have an enormous host range, associating with over 60% of all plant species for the last 460 million years and across the majority of terrestrial biomes (Redecker et al., 2000; Smith and Read, 2008). The fungi improve plant growth, protect plants against pathogens and herbivores, and their diversity has a key role in ecosystem productivity and plant diversity (Harrison, 1997; van der Heijden et al., 1998). Belowground, a single AMF can form extensive networks connecting many different plant species. The mycelium produced by a single AMF can exhibit a diameter of at least 10 m and colonizing different plants (Rosendahl and Stuckenbrock, 2004), resulting in the fungus simultaneously living in different environments. Thirty percent of the total carbon fixed by grassland communities that ends up in the soil is partitioned into the mycorrhizal hyphal network (Drigo et al., 2010).
A major question is how AMF manage to colonize such a wide range of hosts and habitats, both globally and locally. This is especially puzzling because AMF are presumed to be asexual (Judson and Normark, 1996; Sanders and Croll, 2010). Sexual reproduction is thought to be important for allowing organisms to respond to new or changing environments (Weismann, 1889, 1904; Burt, 2000). However, some putatively asexual fungi, such as Candida albicans, have other ways of generating new genotypes in the absence of meiosis (Hickman et al., 2013). How then are AMF able to thrive in such a wide range of conditions when growing vegetatively?
Recent findings could be important for understanding how AMF live in such a diverse set of hosts and environments. Some AMF species are thought to be heterokaryotic, harboring genetically different nuclei (nucleotypes) in a common cytoplasm (Kuhn et al., 2001; Hijri and Sanders, 2005; Angelard et al., 2010). However, there has been no credible explanation regarding the role of heterokaryosis and why it is maintained in AMF (Sanders and Croll, 2010).
The separation of genetic information among different nucleotypes could allow an AMF to undergo rapid genotypic change within its own lifetime in response to environmental change by altering the relative frequencies of different nucleotypes. Such shifts in nucleotype frequencies could potentially allow the fungus to improve its growth in a new environment. It could also allow the fungus to be genotypically different in different parts of the same network. AMF also exhibit partial segregation, producing new vegetative spores with different nucleotype frequencies (Angelard et al., 2010; Ehinger et al., 2012). Thus, an AMF also has a way to produce genotypically different offspring even though they grow vegetatively. These offspring could potentially exhibit a range of different responses to a change of environment compared with their parent, even though they are vegetative offspring. Unlike most organisms that rely on phenotypic plasticity to respond to environmental changes within their lifetimes, changing nucleotype frequencies in AMF potentially confers plasticity to changing environments, more akin to that seen within a population of genotypically different individuals.
The first hypothesis we tested is that a change in environment, specifically a shift in host plant species, will induce changes in the relative frequency of different nucleotypes. This mechanism involves rapid changes in the relative frequency of already existing polymorphism. Genotypic changes correspond in our system to changes in nucleotype frequencies, and, thus, does not require the apparition of new mutations. Our second hypothesis is that genotypic changes due to a shift of host will differ between a parent and offspring produced by partial segregation. On the basis of previous work (Angelard et al., 2010; Angelard and Sanders, 2011), we expect that changes in the relative frequencies of nucleotypes will result in phenotypic changes in the fungi. Thus, our third hypothesis is that because segregated lines are genotypically different from their parents, they will exhibit an overall greater range of phenotypic responses to a host change compared with the initial line. This last hypothesis would indicate that partial segregation in AMF leads to increased plasticity to a change of environment by creating offspring with different phenotypic responses. Testing the second and third hypothesis requires testing for a significant genotype by environment interaction (Stearns, 1992), which in this case would require observing a significantly different genotypic or phenotypic response among segregated lines (genotypes) to a change in environment.
Materials and methods
Cultivation of the AMF Rhizophagus irregularis
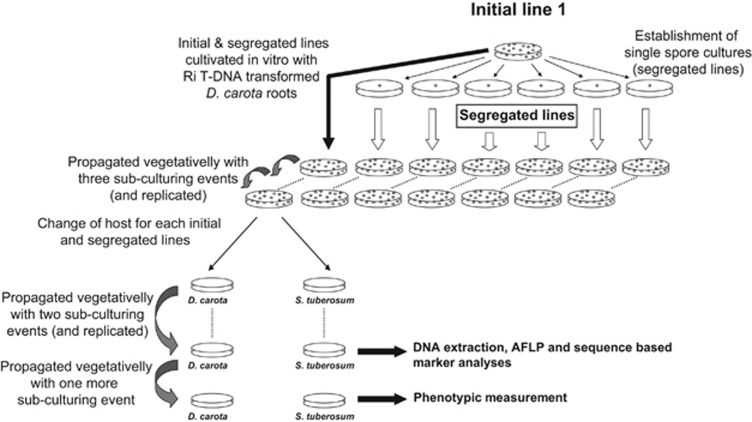
We used an artificial in vitro AMF culture system with transformed roots of different plant species because the system has numerous advantages, such as a precise control of environment in a clean growth system and the possibility to obtain uncontaminated AMF DNA for molecular analyses (Bécard and Piche, 1992; St-Arnaud et al., 1996; Koch et al., 2004). Lines of the AMF R. irregularis, that were previously used to demonstrate segregation, were used in this study (Angelard et al., 2010). These fungal isolates were previously ascribed to the species Glomus intraradices and subsequently as Glomus irregulare (Stockinger et al., 2009). The fungus has, however, recently been renamed as R. irregularis (Krüger et al., 2012). Four initial lines of R. irregularis (S3, S4, Sc1 and Sc2) and three to six segregated lines, initiated from each of the initial lines, were used in the present study (S3a-f, S4a-c, Sc1a-f and Sc2a-f). Segregated lines have been produced by separately cultivating single spores that come from the same parental line. Those segregated lines have been shown to differ genotypically owing to the segregation of nucleotypes at spore formation, a mechanism that has been defined as segregation in Angelard et al. (2010) and that can also be considered as a type of drift. A previous study demonstrated that the phenotypic differences among the segregated lines originating from one parent had a genetic basis (Angelard et al., 2010). Before all current experiments, the segregated lines and their initial lines were cultivated in Petri plates containing root inducing T-DNA-transformed Daucus carota roots (Bécard and Fortin, 1988) and maintained through a minimum of three subculturing events (Figure 1). The AMF lines were subcultured every 15 weeks.
Figure 1.
Conceptual design of the experiment. Segregated lines were started from the isolation and culture of single spores from a given initial line. Each initial and segregated line was then replicated and cultivated under identical conditions with roots of D. carota for 13 months, including three subculturing events (see Materials and Methods). Initial and segregated lines were further maintained on D. carota and also transferred to a new plant host (S. tuberosum). The AMF lines were then cultured for 19 months (including three more subculturing events) on one or the other plant host and the AMF phenotypes and genotypes were measured.
Subculturing AMF lines
Subculturing followed the protocol of Koch et al. (2006). Subculturing AMF lines allows the replication of cultures and maintains genotypic and phenotypic stability among replicates. This involves the transfer of medium containing roots, AMF hyphae and spores to a new plate and does not constitute a new generation. Typically, subculturing starts with 6–12 replicate plates of a given AMF line. The subculturing event involves 6–12 pieces of 2 cm2 of medium being taken from the replicate plates and transferred to 6–12 new plates. All the replicate plates of one subculture event are not necessarily used to establish the next subculture event, several pieces of medium can be taken from one plate. This happens when some plates are contaminated or fail to grow properly, or this is performed to increase the amount of fungal material. See Supplementary Methods for more details about subculturing.
Host shift and measurements of the phenotype
After growth on D. carota roots, initial and segregated AMF lines were used to establish new axenic cultures on D. carota as well as on root clones of another transformed host: Solanum tuberosum (potato). For each segregated line and each host, 8 replicate plates were established, whereas 10 replicate plates were established for each initial line. The protocol used for changing the root host species was the same as in Ehinger et al. (2009). In summary, cluster of spores and hyphae free of plant roots were transferred to new plates, and new roots (either D. carota or S. tuberosum) were then added to the plates. After the first transfer of the AMF lines onto the new host, each AMF line was then maintained through subculturing events. Spore density was measured with the same protocol as in Koch et al. (2006) to quantify phenotypic differences between strains. We measured spore density because it is a trait linked to fungal fitness (Pringle and Taylor, 2002). We established 10 replicate plates for each AMF line and each host three subculturing events after the change of host, resulting in a total of 500 plates for the phenotypic measurement. The measurement was performed 14 weeks after the beginning of the new subculturing event so that plates had the same amount of time to grow.
Preparation of material for molecular analyses
Hyphae and spores produced vegetatively from AMF lines growing on D. carota and S. tuberosum were used, after two subculturing events, to make two independent DNA extractions per AMF line following Ehinger et al. (2009). In summary, pieces of medium containing fungi and roots were taken from replicate plates and transferred to split plates. Those split plates allowed the proliferation of fungi on one side of the plate but not of plant roots. During subculturing, not all the replicate plates of one AMF line were necessarily used to establish split plates. For example, 5 replicate plates can be used to establish 10 split plates. The medium of each split plate containing fungi were then divided in half and transferred to two containers. In that way, one container received half the fungal material of all split plates and the other container received the other half. One DNA extraction was then performed on the fungal material combined in each container, resulting in two DNA extractions per AMF line. Molecular analyses were then performed on these DNA samples.
Molecular methods
To study genotypic differences due to a host shift, we used amplified fragment length polymorphism, AFLP markers because this method allows a large number of loci to be analyzed. In AMF this method is constrained by the level of detection. Rare nucleotypes, not detectable on one host, could become detectable on the other host if they increased in frequency due to the host shift. Reciprocally, abundant nucleotypes on one host could be lost on the other host or could decrease in frequency and pass below the level of detection. For this reason, a sequence-based marker (Bg112) was also used to look for quantitative changes in allele frequency (Angelard et al., 2010; Ehinger et al., 2012).
APLF to detect genotypic differences among AMF lines on different hosts
The protocols to perform and score AFLP were the same as in Croll et al. (2009), except for minor modifications to the primers and scoring method (see Supplementary Methods and Supplementary Table S1). A total of 1200 loci were analyzed per line and each fragment was scored as follows (Table 1 and Supplementary Table S1):
(1;1) For reproducible fragment (fragment in both replicates exceeded 50 relative fluorescence units).
(1;0) For non-reproducible fragment (fragment in only one replicate exceeded 50 relative fluorescence unit).
(0;0) Where fragment was absent in both AFLP replicates but present in at least one replicate of any AMF line.
Table 1. Method used to score AFLP fragments on each host and comparison of the scores between hosts.
| D. carota Score a | S. tuberosum Score a | Comparison of scores between D. carota and S. tuberosum (score for each locus) |
|---|---|---|
| (0;0) (1;1) | (0;0) (1;1) | No AFLP difference between AMF line growing on two different hosts (0) |
| (1;1) (0;0) | (0;0) (1;1) | AFLP differences between AMF lines growing on two different hosts (1) |
| (1;1) | (1;0) | Non-reproducible fragment for at least |
| (0;0) | (1;0) | one host (2) |
| (1;0) | (1;1) | |
| (1;0) | (0;0) | |
| (1;0) | (1;0) |
Abbreviations: AFLP, amplified fragment length polymorphism; AMF, arbuscular mycorrhizal fungi.
Score (1;1) corresponds to reproducible fragment (fragment present in both AFLP replicate), score (1;0) corresponds to non-reproducible fragments (fragment present in only one replicate) and score (0;0) corresponds to fragment absent in both replicates but present in at least one replicate of any AMF line.
We then compared the AFLP profiles for each AMF line on the two hosts to detect AFLP differences and we scored each locus as follows (Table 1):
(0) For a locus with no AFLP differences between hosts.
(1) For a locus with AFLP differences between hosts.
(2) For a locus with at least one non-reproducible fragment.
Sequence-based marker analysis for the detection of quantitative changes in allele frequencies among AMF lines on different hosts
We carried out quantitative analyses on the frequency of alleles at the Bg112 locus in the AMF lines on different hosts as they signify potential changes in relative nucleotype frequency (Angelard et al., 2010; Ehinger et al., 2012). Four alleles of the Bg112 locus co-occur in each AMF line and can be distinguished by length and sequence polymorphism (Angelard et al., 2010). Four alleles have been identified in the crossed lines S3, Sc1 and Sc2 and their subsequent segregated lines (allele 1: 174bp, allele 2: 178bp, allele 3: 208bp and allele 4: 214bp; accession numbers on GenBank: GU930826, GU930824, GU930827 and GU930828, respectively). Four other alleles have been identified in the crossed line S4 and the segregated lines S4a, S4b and S4c (allele 5: 160bp, allele 6: 168bp, allele 7: 174bp and allele 8: 180bp; accession numbers on GenBank: GU930825, GU930836, GU930835 and GU930834, respectively). We replicated the PCR reactions twice, each with 5 ng of DNA as the starting template. Four values were then obtained for each allele and each AMF line (two PCR reactions on each of the two independent DNA extractions). We used a protocol previously described for the PCR reactions (Croll et al., 2009), except that the number of PCR cycles was adapted. The relative frequency of each allele was measured as in Angelard et al. (2010). See Supplementary Methods for more details on the number of values obtained for each AMF line and on the calculation of allele frequency.
Pyrosequencing to detect quantitative changes in the frequency of alleles among AMF lines on different hosts
We also looked for quantitative changes in allele frequency among AMF lines by pyrosequencing of amplicons of the Bg112 locus (Angelard et al., 2010). Four independent replicates were generated for each AMF line on each host (two PCR reactions on each of the two independent DNA extractions). The analyses were performed with DNA of the initial line S4 and the segregated lines S4b and S4c. Four Bg112 alleles were detected (alleles VI–IX). The DNA sequences are archived at NCBI SRA: SRP028250. More details are available in the Supplementary Methods.
Statistical analyses
Genotypic changes observed by AFLP
1/Test assessing whether AFLP differences observed between hosts can be explained without an effect of host change. We performed AFLP on two independent DNA extractions, giving two AFLP profiles per AMF line. Therefore, the detection of a non-reproducible fragment (1;0) could arise from three sources of variation: (a) a rare allele with frequency around the threshold detection level of AFLP method; (b) by mutation(s) in one replicate, increasing sufficiently in frequency to become detectable and finally; (c) methodological errors (such as technical errors or differences in DNA quality).
The AFLP differences observed between hosts could be due to the same three sources of variation described above. In addition, a shift of host could induce a plastic response and modify the frequency of alleles. In others words, more variation between hosts than between replicates would suggest an effect of a change of host on the genotypic content of a particular AMF line. In order to test this hypothesis, we used a probability framework (described in Supplementary Methods) to quantify the variation between replicates with an error rate Er that encapsulates the three sources of variation between replicates described above, to predict the expected variation between hosts without host effect (Pdiff) and to compare it with the observed variation between hosts (Pobs).
We also then performed a Fisher's Omnibus test on the whole data set as it integrates the individual P-values for the repeated tests into one test that summarizes whether there was an overall host effect in the AFLP data set.
2/Comparison of the number of AFLP differences among AMF lines. After having tested whether AFLP differences found for each AMF line could not be explained without an effect of change of host, we compared the number of AFLP differences among AMF lines. We first performed a global G-test of independence (Sokal and Rohlf, 1995) to compare the overall number of AFLP differences. We then performed four G-tests of independence with the Holm's correction for multiple tests (one test for each initial line and its respective segregated lines) to compare the number of AFLP differences among AMF lines. Finally, we performed four one-sided proportion tests (Newcombe, 1998), with the Holm's correction for multiple tests, to determine whether the proportion of AFLP changes in the initial line was lower than the proportion of AFLP changes in the pooled segregated lines.
3/Permutation test to determine whether AFLP differences due to host change are consistent across AMF lines or display random patterns. We considered the total number of AFLP differences found for all the AMF lines. The goal of the test was to determine whether these AFLP differences were randomly distributed among the 1200 loci analyzed in this study. We did this because test 1 (described above) involves repeated tests for a host effect on each AMF line but with very low replication per test. Each individual test yields a significant effect that would be unlikely to happen repeatedly by chance. A permutation test (Manly, 2007) on the AFLP data allowed us to see whether a change of host consistently and significantly affected a group of loci in a non-random way. To do this, we compared the observed ratio of ‘total number of loci with AFLP differences/total number of AFLP differences' with the ratios of 1000 permuted replicates where AFLP differences permutations were constrained within AMF lines. The P-value in this test represents the probability of obtaining the observed changes in loci by chance. See Supplementary Methods for more details.
Test for quantitative changes in allele frequency observed in the Bg112 locus
To detect quantitative changes due to a host shift, multivariate analyses of variance were performed on the relative peak heights of the four alleles. Two-way multivariate analyses of variance with the main factors ‘AMF line' and ‘host species' were performed, one for each initial line (and their subsequent segregated lines).
Host effects on AMF phenotypes
We first tested the overall effect of a change of host on the growth of the AMF lines on the whole data set. To do this, we grouped each initial AMF line and its segregated lines together, resulting in four ‘initial AMF' categories, one for each initial AMF (for example ‘initial AMF' S3 regroups all the lines coming from S3, including S3). Because of the repeated measures of AMF lines in our experimental design, we used a linear mixed-effects model (Pinheiro and Bates, 2000; Bates and Sarkar, 2007) in which ‘initial AMF', ‘host species' and the ‘initial AMF:host species' interaction were held as fixed effects, and AMF lines (segregated and initial) nested within its ‘initial AMF' was included as a random effect. Main effects were analyzed using analysis of variance with marginal sums of squares (Fox, 2008; Fox and Weisberg, 2011). We used cubed-root transformed values of spore density to meet model assumptions of normality.
Because we observed an initial AMF effect and an initial AMF by host interaction, we then analyzed the initial AMF separately. For each initial AMF, we used a linear mixed-effects model in which ‘AMF line', ‘host species' and the ‘AMF line:host species' interaction were held as fixed effects, and variance among AMF lines was included as a random effect.
All the analyses were performed using the program JMP version 5.0 (SAS Insitute Inc, Cary, NC, USA), R version 2.0.1 (R development Core Team, 2003, www.R-project.org) and Mathematica version 7.0 (Wolfram Research Inc, Champaign, IL, USA; 2008).
Results
Genotypic changes at multiple loci due to a host shift
1/Test assessing whether AFLP differences observed between hosts can be explained without an effect of host change
The test showed that the three sources of variation (that is, limits of detection, mutation and experimental errors) were not enough to explain the observed differences in AFLP patterns between hosts in the whole data set (Fisher's Omnibus: χ2=81.4, degree of freedom=46, P<0.001). This was also the case for all the AMF lines analyzed separately (Table 2). Thus, the data indicate in repeated independent tests that AMF lines growing on potato had diverged from those growing on carrot. Therefore, genotypic divergences owing to a shift of host must have occurred within all AMF lines. The proportion of non-reproducible markers was very low compared with the number of reproducible markers, showing that the AFLP technique was a reliable method for the analysis of host-induced changes (Supplementary Table S1).
Table 2. Number of AFLP differences and results of the test assessing whether AFLP differences observed between hosts can be explained without an effect of host change.
| AMF lines | Number of AFLP differences | Error rate (Er) |
Comparison of AFLP differences between hosts
|
|
|---|---|---|---|---|
| Expected proportion (Pdiff) | Observed proportion (Pobs) | |||
| S3 | 33 | 0.011 | 0.0002 | 0.0275a |
| S3a | 65 | 0.025 | 0.0012 | 0.0542a |
| S3b | 43 | 0.012 | 0.0003 | 0.0358a |
| S3c | 43 | 0.007 | 0.0001 | 0.0358a |
| S3d | 51 | 0.008 | 0.0001 | 0.0425a |
| S3e | 52 | 0.027 | 0.0013 | 0.0433a |
| S3f | 40 | 0.016 | 0.0005 | 0.0333a |
| S4 | 14 | 0.012 | 0.0003 | 0.0117a |
| S4a | 30 | 0.011 | 0.0003 | 0.0250a |
| S4b | 50 | 0.005 | 0.0001 | 0.0417a |
| S4c | 30 | 0.004 | 0.0000 | 0.0250a |
| Sc1 | 18 | 0.018 | 0.0006 | 0.0150a |
| Sc1a | 42 | 0.010 | 0.0002 | 0.0350a |
| Sc1b | 34 | 0.018 | 0.0006 | 0.0283a |
| Sc1c | 41 | 0.014 | 0.0004 | 0.0342a |
| Sc1d | 20 | 0.015 | 0.0005 | 0.0167a |
| Sc1e | 22 | 0.026 | 0.0013 | 0.0183a |
| Sc1f | 45 | 0.008 | 0.0001 | 0.0375a |
| Sc2 | 31 | 0.021 | 0.0008 | 0.0258a |
| Sc2c | 30 | 0.008 | 0.0001 | 0.0250a |
| Sc2d | 44 | 0.014 | 0.0004 | 0.0367a |
| Sc2e | 26 | 0.007 | 0.0001 | 0.0217a |
| Sc2f | 27 | 0.011 | 0.0002 | 0.0225a |
Abbreviations: AFLP, amplified fragment length polymorphism; AMF, arbuscular mycorrhizal fungi.
The probability of Pobs>Pdiff is lower than 0.05, in which case we consider that genotypic divergence due to a shift of host must have occurred within an AMF line.
2/Comparison of the number of AFLP differences
Overall, we found that the number of AFLP differences, owing to a host shift, were not the same among all AMF lines (Global G-test: G22=99.8, P<0.001). Because in almost every case, the proportions of AFLP differences due to a change of host were higher in segregated lines than in their respective initial lines (Table 2), we then compared the number of AFLP differences for each initial line and its segregated lines. In each case, we found a significant difference among AMF lines (Table 3). Finally, we found that the proportions of AFLP changes were significantly lower overall in initial versus segregated lines (Fisher's Omnibus on corrected P-values: χ2=29.95, degree of freedom=8, P<0.001) and for three out of four initial lines than in their respective segregated lines (Table 3).
Table 3. Results of the G-tests comparing the number of AFLP differences among AMF lines, and results of the proportions tests comparing the proportion of AFLP differences between initial and segregated lines.
| G-test | Proportion test | |||
|---|---|---|---|---|
| Initial line S3 and respective segregated lines | G6=26.2 | P<0.001 |
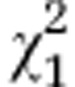 =4.45 =4.45 |
P=0.035 |
| Initial line S4 and respective segregated lines | G3=29.7 | P<0.001 |
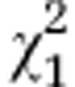 =12.02 =12.02 |
P=0.001 |
| Initial line Sc1 and respective segregated lines | G6=39.2 | P<0.001 |
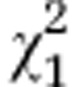 =6.60 =6.60 |
P=0.015 |
| Initial line Sc2 and respective segregated lines | G6=12.5 | P=0.014 |
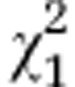 =4e-4 =4e-4
|
P=0.492 |
Abbreviations: AFLP, amplified fragment length polymorphism; AMF, arbuscular mycorrhizal fungi.
3/Permutation test
The number of loci with AFLP differences was retained within a smaller set of loci (observed ratio=0.187) than expected by chance (P<0.001). This means that the subset of loci that were affected by a host change were not just a random sample of loci but that similar loci were affected by a host change among AMF lines. In other words, the majority of AFLP polymorphism was found at the same loci among AMF lines.
Quantitative changes in nucleotype frequency due to a host shift
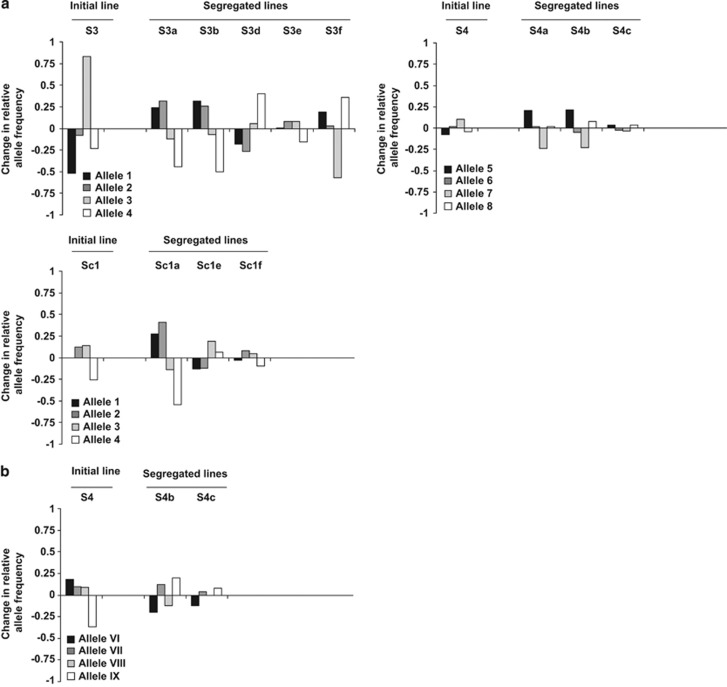
The same four alleles were found in the initial lines S3 and Sc1 and their segregated lines (alleles 1–4). Another four alleles were found in the initial line S4 and the segregated lines S4a-c (alleles 5–8). We found a significant AMF line by host interaction for all the lines, meaning that some alleles had significantly increased in frequency, whereas another allele (or alleles) had significantly decreased in frequency, and those changes were not the same across AMF lines (Figure 2a; Table 4 and Supplementary Table S2). For example, allele 1 decreased in frequency in response to a change of host in line S3 and S3d, whereas the same allele increased in frequency in S3a, S3b and S3f in response to the change of host (Figure 2a). As a confirmation of our results, 454 pyrosequencing data showed a significant alteration in relative allele frequencies among the lines (Figure 2b; Table 4 and Supplementary Table S2).
Figure 2.
Relative frequency of four Bg112 alleles was altered by a host shift. Change in the relative frequency of Bg112 alleles following a host shift. Positive values represent an increase in frequency of an allele due to a shift from D. carota to S. tuberosum and negative values represent a decrease in the frequency of a given allele due to a shift from D. carota to S. tuberosum. (a) Change in frequency of alleles measured on three initial lines and their segregated lines. (b) Change in frequency of alleles for line S4 and its segregated lines as measured by 454 pyrosequencing. See Table 4 for associated multivariate analysis of variance (MANOVA) statistics.
Table 4. Multivariate analysis of variance (MANOVA) results on the relative frequency of alleles of the locus Bg112 following a host shift from D. carota to S. tuberosum. Numbers in parentheses represent the Pillai Trace value.
| AMF lines | Host | AMF lines:host | |
|---|---|---|---|
| Analysis using capilliary electorphoresis | |||
| Initial line S3 | F20,160=2.46 (0.94)* | F4,32=1.07ns | F20,160=4.04 (1.34)*** |
| Initial line S4 | F12,75=2.41 (0.84)* | F4,23=1.48ns | F12,75=2.46 (0.86)** |
| Initial line Sc1 | F12,75=1.16 (0.47)ns | F4.22=5.00** | F12,75=1.16 (0.47)* |
| Analysis using 454 pyrosequencing | |||
| Initial line S4 | F8,32=7.26 (1.29)*** | F4,15=8.13*** | F8,32=5.19 (1.13)*** |
Abbreviations: AMF, arbuscular mycorrhizal fungi; ns, not significant.
Significance levels are: ***, P<0.001; **, P<0.01; *, P<0.05; ns, P>0.05.
Effects of host shift and segregation on AMF fitness
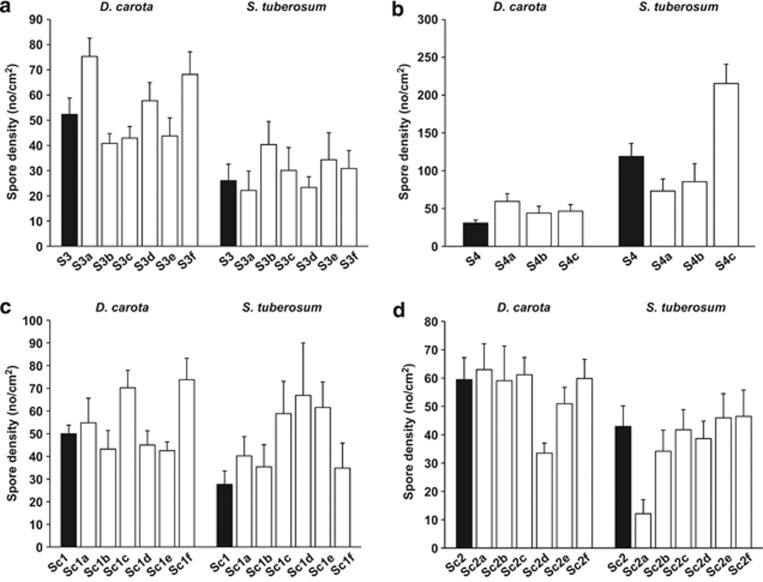
In the whole data set, we found that both ‘initial AMF' and ‘host species' had significant effects on fungal spore density (Table 5a). Furthermore, there was a strong interaction between those two factors, indicating that each set of ‘initial AMF' differed in their response to a change of host (Table 5a, Figure 3). Because ‘initial AMF' differed in their response to host shift (‘initial AMF:host species' interaction; Table 5a), we also analyzed the initial AMF separately (Table 5b). The factor ‘AMF line' is not informative in these analyses because we defined AMF lines (segregated and initial) nested within its ‘initial AMF' as a random effect. However, among the four initial AMF after controlling for the variance among AMF lines, we found significant ‘host species:AMF line' interactions on spore density for all the lines as well as significant ‘host species' effects for three of the four initial AMF (Table 5b). In some lines, a host shift did not confer fitness gains to the AMF lines. However, in several cases segregation produced a variety of lines, some of which achieved a significantly greater fitness on the new host compared to the old host (Figure 3).
Table 5. Results of the linear mixed-effects model testing the effect of a change of host on overall AMF growth (A) and on the growth of each initial line and their segregated lines (B).
| (A) | Initial AMF | Host | Initial AMF:host |
|---|---|---|---|
| Overall effect |
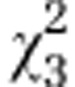 =10.95* =10.95* |
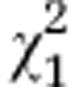 =11.99*** =11.99*** |
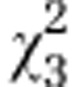 =94.09*** =94.09*** |
| (B) |
AMF line
|
Host
|
AMF line:host
|
| Initial line S3 |
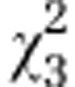 =0.44ns =0.44ns
|
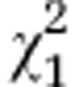 =44.21*** =44.21*** |
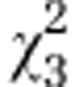 =20.42** =20.42** |
| Initial line S4 |
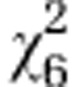 =3.96ns =3.96ns
|
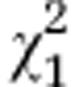 =43.78*** =43.78*** |
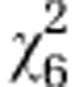 =20.75*** =20.75*** |
| Initial line Sc1 |
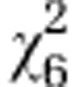 =2.83ns =2.83ns
|
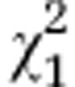 =2.94ns =2.94ns
|
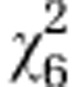 =15.91* =15.91* |
| Initial line Sc2 |
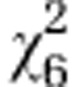 =5.68ns =5.68ns
|
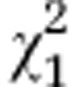 =26.42*** =26.42*** |
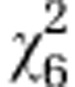 =30.81*** =30.81*** |
Abbreviations: AMF, arbuscular mycorrhizal fungi; ns, not significant.
Significance levels are: ***, P<0.001; **, P<0.01; *, P<0.05; ns, P>0.05.
Figure 3.
Host shift altered phenotypes of the AMF lines. Spore of initial and segregated lines growing on two hosts D. carota and S. tuberosum. (a) Line S3 and its segregated lines. (b) Line S4 and its segregated lines. (c) Line Sc1 and its segregated lines. (d) Line Sc2 and its segregated lines. Black bars: initial lines. White bars: segregated lines. See Table 5 for statistical analyses.
Discussion
Large and rapid genotypic plasticity in response to host shift
We used three molecular techniques showing that a change of host resulted in genotypic changes in AMF within the lifetime of the fungus. These changes were consistent and did not randomly affect different loci among the lines but significantly affected the same combination of loci. AFLP, normally yields qualitative data about allele presence and absence. However, in AMF, the results may reflect quantitative changes in allele frequency. The percentage of genotypic changes as shown by AFLP appears high (1.17–5.42% Table 2) given that this represented a short period of vegetative growth. These figures should not be interpreted as a true loss or gain of alleles as they likely reflect changes in nucleotype frequency. These data support the first hypothesis, that AMF respond rapidly to a change of environment through changes in the frequency of nucleotypes, and confirm the following: (1) a host shift significantly alters the genotype of AMF in a qualitative or quantitative way (Tables 2, 3); (2) the change involves large numbers of alleles (Table 2); and (3) the change is rapid. The data also showed that the amount of genotypic change due to a change of host was significantly different among each initial line and their segregated lines (Table 3), which was the second hypothesis. Moreover, the genotypic changes due to host change in initial lines were in almost all cases smaller than in segregated lines, suggesting that segregated AMF lines were more strongly altered genotypically (by changes in nucleotype frequencies) by the host shift than were their initial lines (Tables 2, 3). Because some rare nucleotypes could be lost during the formation of the segregated lines, we expected nucleotype diversity to be higher in initial lines than in segregated lines. Consequently, we expected that a greater effect of host change would be seen in initial lines than in segregated lines, but this was not the case. This highlights the importance of segregation for genotypic plasticity in these asexual fungi.
The results of the quantitative molecular analyses showed that a host shift induced a significant alteration in relative allele frequencies among initial and segregated AMF lines. This result supports the first and second hypothesis. This result shows that alleles were likely to be located on different nuclei inside the fungus and that the different nuclei had changed frequency in response to the host shift. If all four alleles had been colocated on each nucleus then an increase in the frequency of one allele due to a host shift would have been correlated with an increase in the frequency of another allele. This cannot be the case if a significant AMF line by host interaction on relative allele frequencies is observed using a multivariate analysis of variance because this means that the frequency of the alleles responded significantly differently from each other to a change of host (Table 4). This is the only plausible explanation that we can propose to explain this data.
Our work suggests that AMF are genotypically plastic in response to the environment that they experience, and that this is rapidly manifested by changes in nucleotype frequencies within the fungus. We consider this as a mechanism by which AMF can respond genotypically (in a rapid manner) to the environment in a way that is analogous to a population of genetically different individuals responding to a change in environment. In numerous studies, genetic variation has been shown to be a key factor in the ability of a population to respond to environmental change. This is because not all genotypes in a population have the level of plasticity that allows them to do well in all environments (Stearns, 1992). Our work suggests that fluctuations in nucleotype frequency in response to a host change potentially provide a larger degree of plasticity than that which could be achieved if the fungus contained one nucleotype. Given that it is known that alteration in nucleotype frequency in AMF alters the phenotype, such effects have the potential to increase AMF phenotypic plasticity to the environments they experience. This study assumes clonality in AMF. The results would, thus, indicate that even if AMF are clonal, mechanisms exist to enhance their plasticity in changing environments. However, it should be noted that a full complement of genes necessary for meiosis exist in the genome of AMF and that signatures of recombination have been observed in AMF populations (Croll and Sanders, 2009; Halary et al., 2011), although it is impossible to know whether meiosis occurs and, if so, whether it is frequent. Even if AMF undergo some meiosis, this would not invalidate the interesting ecological effects of plasticity observed in this study in response to a host shift.
Phenotypic plasticity in response to host change is enhanced by segregation
The fungal growth results support the third hypothesis that segregation produced a variety of genotypically different AMF lines that displayed a wide range of fitness to the new host and that differed from the initial lines. Our results suggest that segregation increased the ability of the fungus to create new individuals with a wider range of phenotypic responses to the environment, indicating increased plasticity. Anastomosis has been observed in AMF and this would allow the remixing of nuclei following segregation (Sanders and Croll, 2010). However, whether anastomosis occurs in nature, how often anastomosis occurs in nature, how much genetic material is transferred from one individual to another and the proportion of those nuclei that survive and replicate following anastomosis is completely unknown. Without this information, it is impossible to say how much the effects of anastomosis offset segregation effects in nature. The host effect on the fungal genotype also happened in non-segregated lines and, thus, should also act on anastomosed lines. Thus, frequent anastomosis would not invalidate our results.
Are the genotypic changes in AMF in response to a host adaptive?
That nucleotype frequency was altered by a change in environment shows that some nuclei were favored over others by a change of environment; a prerequisite for adaptation. Despite the fact that the total amount of genotypic changes due to a change of environment varied among AMF lines, we also showed that those genotypic changes did not occur randomly. Instead, we showed that similar genotypic changes happened within each AMF line in term of nucleotype composition, thus indicating common genotypic responses to the change of environment. Moreover, we also found an overall effect of the change of host on the growth of the fungi, as well as a significant overall interaction between AMF lines and host species. Those results show that the plant host has an effect on the production of spores by AMF and that AMF are not responding in the same way to different plant hosts.
We cannot conclude from these data that a host-induced change in nucleotype frequency directly caused the observed change in the fitness of the fungus in the new host. However, adaptation occurs in a new environment when selection acts on genetic variation, favoring some genotypes over others. Thus, our results show the potential for adaptation to occur within an AMF in its lifetime owing to its genetic organization.
The design of this experiment, however, allows us to draw a link between changes in nucleotype frequency and the change in fitness of the fungus in response to a host change. Segregated and initial lines differed in their nucleotype frequencies. The phenotypic responses among replicate cultures of a segregated line showed little variation compared with that among segregated lines (a significant genotype × environment interaction; Table 5). If the phenotypic responses to a change of host were only epigenetic then more variation in phenotypes among replicates of the same segregated line would be expected. Therefore, the design (and replication) in this experiment allows us to conclude that the difference in phenotypic response to host change among segregated lines has a genetic basis that involves changes in nucleotype frequency. This indicates that changes in nucleotype frequency have the potential to be adaptive. Some segregated lines appeared to be maladapted to the new host as some had a lower fitness in the new environment. However, in nature, AMF encounter many different host species, and a basic premise of evolutionary ecology is that one genotype cannot have optimal fitness in all environments (Stearns, 1992). Thus, AMF lines that exhibited low fitness on potato could have a higher fitness on another host species that they may encounter.
Ecological relevance and conclusions
Our results demonstrate that AMF can rapidly undergo genotypic change in response to the environment they experience, and that changes in nucleotype frequency can have a role in performance in a new environment. In addition, the segregation of nucleotypes among AMF lines leads to novel AMF offspring, even though they grow vegetatively, that display greater genotypic change and plasticity than their parent in response to a host shift. Overall, our results have several ecologically relevant points. First, the ability to rapidly change genotype composition is important as it provides a mechanism for the fungus to be plastic when growing from plant to plant. AMF networks make up a large amount of belowground microbial biomass, which must be due to the fact that a fungus can colonize so many different plants. It is, therefore, essential to understand how AMF do this. Second, the ability to alter nucleotype frequencies offers a greater potential for adaptability to the different environments the fungus experiences than that which could occur by plasticity in one genome. The potential magnitude of this is evident given the large number of alleles affected by a host change. Third, our results mean that AMF have the capacity to be genotypically different in the hyphal network when forming symbioses simultaneously with different plant species. Thus, our results highlight an ecological advantage to AMF afforded by separation of genetic variation among different nuclei.
Finally, the link between differences in nucleotype frequency and fitness changes in response to host change strongly point to segregation as a potential mechanism in these fungi to increase plasticity to new environments. This is the only study that we know of that highlights the role of AMF genetics as a potential mechanism for wide plasticity and adaptation of AMF to changing environments. Understanding how these abundant and ecologically important components of terrestrial ecosystems react to rapid environmental change may help to avoid adverse outcomes in the future (Johnson et al., 2013). Consequently, we hope this work will stimulate research in this area.
Acknowledgments
We thank M Bucher (University of Cologne) and G Bécard (University Paul Sabatier, Toulouse) for supplying roots and Jeremy Bonvin for help with DNA extraction and molecular analyses. We also thank T Kiers, T Kawecki and J Goudet for constructive comments on this work. This work was supported by a grant from the Swiss National Science Foundation (310030B_144079/1). Correspondence and request for materials should be addressed to IRS (ian.sanders@unil.ch).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Angelard C, Colard A, Niculita-Hirzel H, Croll D, Sanders IR. Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr Biol. 2010;20:1216–1221. doi: 10.1016/j.cub.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Angelard C, Sanders IR. Effect of segregation and genetic exchange on arbuscular mycorrhizal fungi in colonization of roots. New Phytol. 2011;189:652–657. doi: 10.1111/j.1469-8137.2010.03602.x. [DOI] [PubMed] [Google Scholar]
- Bates DM, Sarkar D.2007. Lme4: Linear mixed-effects models using S4 classes. R package version 0.999999-0.
- Bécard G, Fortin JA. Early events of vesicular arbuscular mycorrhiza formation on Ri T-DNA trnasformed roots. New Phytol. 1988;108:211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x. [DOI] [PubMed] [Google Scholar]
- Bécard G, Piche Y. Establishment of vesicular-arbuscular mycorrhiza in root organ culture - Review and proposed methodology. Methods Microbiol. 1992;24:89–108. [Google Scholar]
- Burt A. Sex, recombination and the efficacy of selection - was Weissman right. Evolution. 2000;54:337–351. doi: 10.1111/j.0014-3820.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Croll D, Sanders IR. Recombination in Glomus intraradices, a supposed ancient asexual arbuscular mycorrhizal fungus. BMC Evol Biol. 2009;9:13. doi: 10.1186/1471-2148-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Giovannetti M, Koch AM, Sbrana C, Ehinger M, Lammers PJ, et al. Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2009;181:924–937. doi: 10.1111/j.1469-8137.2008.02726.x. [DOI] [PubMed] [Google Scholar]
- Drigo B, Pijl AS, Duyts H, Kielak A, Gamper HA, Houtekamer MJ, et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Nat Acad Sci. 2010;107:10938–10942. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehinger M, Croll D, Koch MA, Sanders IR. Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytol. 2012;196:853–861. doi: 10.1111/j.1469-8137.2012.04278.x. [DOI] [PubMed] [Google Scholar]
- Ehinger M, Koch AM, Sanders IR. Changes in arbuscular mycorrhizal fungal phenotypes and genotypes in response to plant species identity and phosphorus concentration. New Phytol. 2009;184:412–423. doi: 10.1111/j.1469-8137.2009.02983.x. [DOI] [PubMed] [Google Scholar]
- Fox J.2008Applied Regression analysis and Generalized Linear Models2nd edn.Sage Publications: Thousand Oaks, CA, USA [Google Scholar]
- Fox J, Weisberg S.2011An R companion to applied regression2nd edn.Sage Publications: Thousand Oaks, CA, USA [Google Scholar]
- Halary S, Malik S-B, Lildhar L, Slamovits CH, Hijri M, Corradi N. Conserved meiotic machinery in Glomus spp., a putatively ancient asexual fungal lineage. Genome Biol Evol. 2011;3:950–958. doi: 10.1093/gbe/evr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ. The arbuscular mycorrhizal symbiosis: An underground association. Trends Plant Sci. 1997;2:54–60. [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, et al. The ‘obligate diploid' Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijri M, Sanders IR. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature. 2005;433:160–163. doi: 10.1038/nature03069. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Angelard C, Sanders IR, Kiers ET. Predicting community and ecosystem outcomes of mycorrhizal responses to global change. Ecol Lett. 2013;16:140–153. doi: 10.1111/ele.12085. [DOI] [PubMed] [Google Scholar]
- Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996;11:41–46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- Koch AM, Croll D, Sanders IR. Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol Lett. 2006;9:103–110. doi: 10.1111/j.1461-0248.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- Koch AM, Kuhn G, Fontanillas P, Fumagalli L, Goudet J, Sanders IR. High genetic variability and low local diversity in a population of arbuscular mycorrhizal fungi. Proc Nat Acad Sci USA. 2004;101:2369–2374. doi: 10.1073/pnas.0306441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M, Krüger C, Walker C, Stockinger H, Schussler A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol. 2012;193:970–984. doi: 10.1111/j.1469-8137.2011.03962.x. [DOI] [PubMed] [Google Scholar]
- Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414:745–748. doi: 10.1038/414745a. [DOI] [PubMed] [Google Scholar]
- Manly BFJ.2007Randomization, Bootstrap, and Monte Carlo Methods in Biology3rd edn.Chapman and Hall: London, UK [Google Scholar]
- Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med. 1998;17:873–890. doi: 10.1002/(sici)1097-0258(19980430)17:8<873::aid-sim779>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. Springer: New York, NY, USA; 2000. [Google Scholar]
- Pringle A, Taylor JW. The fitness of filamentous fungi. Trends Microbiol. 2002;10:474–481. doi: 10.1016/s0966-842x(02)02447-2. [DOI] [PubMed] [Google Scholar]
- Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289:1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- Rosendahl S, Stuckenbrock EH. Community structure of arbuscular mycorrhizal fungi in undisturbed vegetation revealed by analyses of LSU rDNA sequences. Mol Ecol. 2004;13:3179–3186. doi: 10.1111/j.1365-294X.2004.02295.x. [DOI] [PubMed] [Google Scholar]
- Sanders IR, Croll D. Arbuscular mycorrhiza: The challenge to understand the genetics of the fungal partner. Annu Rev Genet. 2010;44:271–292. doi: 10.1146/annurev-genet-102108-134239. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. Academic Press: San Diego, CA, USA; 2008. [Google Scholar]
- Sokal RR, Rohlf FJ.1995Biometry: The principles and practice of statistics in biological research3rd edn.W.H. Freeman and Co: New York, NY, USA [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res. 1996;100:328–332. [Google Scholar]
- Stearns SC. The Evolution of Life Histories. Oxford University Press: Oxford, UK; 1992. [Google Scholar]
- Stockinger H, Walker C, Schussler A. ‘Glomus intraradices DAOM197198', a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 2009;183:1176–1187. doi: 10.1111/j.1469-8137.2009.02874.x. [DOI] [PubMed] [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- Weismann A.1889The significance of sexual reproduction in the theory of natural selectionIn: SSEB Poulton, Shipley AE (eds)Essays upon heredity and kindred biological problems Clarendon Press: Oxford, UK [Google Scholar]
- Weismann A. The Evolution Theory. Edward Arnold: London, UK; 1904. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.