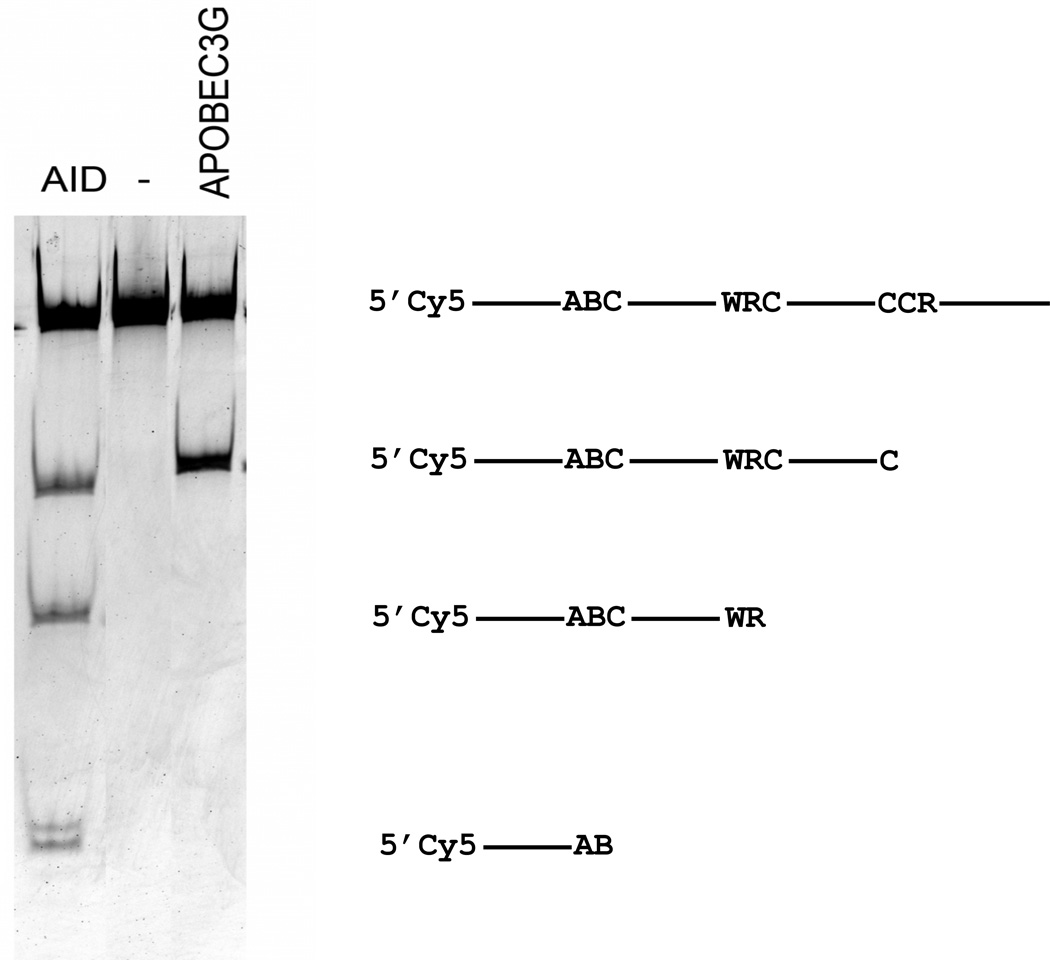
Fig. 4.
Preference of hAID and hAPOBEC3G for specific motifs as judged by in vitro deamination assay. Activity of the two enzymes was compared with the use of Cy5 labeled oligonucleotide containing three sequence motifs (“Materials and Methods”) matching hotspot motifs for cytosine deamination (Table 3). The gel image to the left is the result of separation of processed deamination reaction products on denaturing polyacrylamide gel. The diagram to the right explains what sites were deaminated to generate the observed bands.