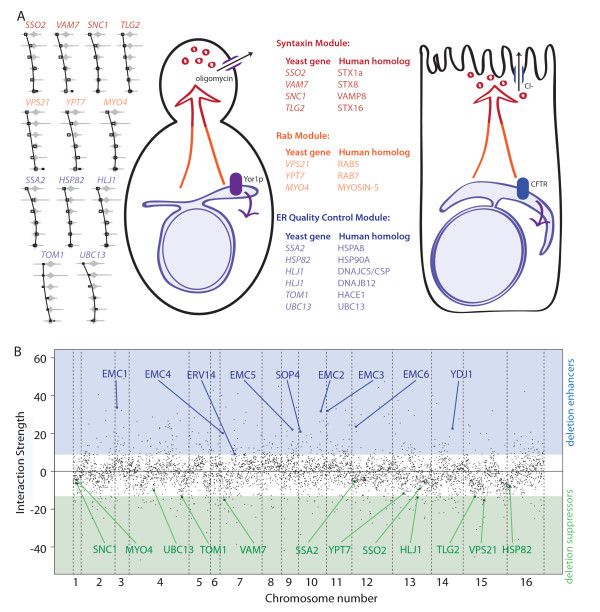
Figure 5.
Modulation of CFTR-ΔF and Yor1-ΔF biogenesis occurs via homologous gene interaction. (A) At left, gene interactions are illustrated by plotting the parameter L vs. oligomycin concentration (increasing upward: 0, 0.05, 0.1, 0.15, 0.2, and 0.25 µg/mL) for the double mutant and the full distribution of 768 Yor1-ΔF single mutant cultures (gray diamonds). Leftward departure indicates faster growth and improved biogenesis of Yor1-ΔF. At right, human genes found in the literature to modulate CFTR-ΔF biogenesis are paired with homologous yeast genes. Gene pairs are grouped into protein classes associated with discrete cellular functions (see text and Additional File 1 - Discussion C). (B) The strength of gene interaction is depicted with respect to chromosomal position and highlighted gene interactions indicate those discussed in the manuscript (interactions are calculated at an oligomycin concentration of 0.2 µg/mL). Vertical lines demarcate chromosomes.