Figure 6.
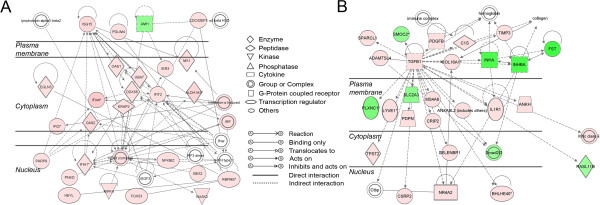
The top two networks (A and B) determined in IPA. Genes were differentially regulated between TNFα (± FSH)-treated and control (± FSH)-treated granulosa cells (n = 379 genes mapped to IPA). The networks were generated by a modified triangle connectivity algorithm based on the proportion of known interactions between molecules in this dataset (focus molecules) and others in the IPA database. Both networks were scored at 44 (−log of P value of Fisher’s exact t-test), and mapped 26 genes each from the differentially expressed dataset. Interactions between molecules are shown as explained in the legend, with focus molecule symbols highlighted in colour, based on up (red) or down (green) regulation and increasing colour intensity with degree of fold change.
