Abstract
In sexually reproducing animals, primordial germ cells (PGCs) are often set aside early in embryogenesis, a strategy that minimizes the risk of genomic damage associated with replication and mitosis during the cell cycle. Here, we have used germ line markers (piwi, vasa, and nanos) and microinjected cell lineage tracers to show that PGC specification in the leech genus Helobdella follows a different scenario: in this hermaphrodite, the male and female PGCs segregate from somatic lineages only after more than 20 rounds of zygotic mitosis; the male and female PGCs share the same (mesodermal) cell lineage for 19 rounds of zygotic mitosis. Moreover, while all three markers are expressed in both male and female reproductive tissues of the adult, they are expressed differentially between the male and female PGCs of the developing embryo: piwi and vasa are expressed preferentially in female PGCs at a time when nanos is expressed preferentially in male PGCs. A priori, the delayed segregation of male and female PGCs from somatic tissues and from one another increases the probability of mutations affecting both male and female PGCs of a given individual. We speculate that this suite of features, combined with a capacity for self-fertilization, may contribute to the dramatically rearranged genome of Helobdella robusta relative to other animals.
Introduction
The near ubiquity of sexual reproduction among modern animal taxa and the involvement of conserved sets of genes in specifying their primordial germ cells (PGCs) indicate that sexual reproduction via specialized germ cells is ancestral to Bilateria. Conserved genes involved in PGC specification include homologs of vasa, piwi, and nanos. Vasa is an ATP-dependent RNA DEAD box helicase that plays an essential role in the establishment and maintenance of the germ line by promoting the translation of germline-specific mRNAs (Carrera et al. 2000; Fujimura and Takamura 2000; Komiya et al. 1994). Piwi, characterized by an RNAase H domain, is involved in cell autonomous, self-renewing, germ stem cell maintenance (Cerutti et al. 2000; Cox et al. 2000; Song et al. 2004; Seto et al. 2007). Nanos is a translational repressor characterized by a C-terminal domain comprising two CCHC zinc-finger motifs (Wang and Lin 2004; Ewen-Campen et al. 2010).
Previous work has identified two distinct modes of germ cell specification, with names rooted in old debates concerning the nature of development itself: preformation entails the segregation of germ line determinants during early embryogenesis while epigenesis entails induction of the germ line at later stages by signals from neighboring cells (Extavour and Akam 2003). Comparative studies suggest that preformation and epigenesis are not mutually exclusive. For example, the proposed model of germ line specification for the polychaete annelid Platynereis dumerilii entails a combination of inherited cytoplasmic determinants, which establish a mesodermal posterior growth zone (MPGZ), followed by inductive processes to determine which cells of the MPGZ become the PGCs (Rebscher et al. 2007). Related findings in diverse taxa have led to the hypothesis that a broadly conserved germline multipotency program (GMP; involving Piwi, Nanos, Vasa, and other gene products) distinguishes a class of primordial stem cells (PriSCs), which are intermediate between the zygote and the PGCs, and which also give rise to somatic cells (Juliano et al. 2010; Solana 2013).
Leeches of the genus Helobdella (Annelida: Clitellata: Hirudinea: Glossiphoniidae) provide another annelid model that is evolutionarily distant from Platynereis (Struck et al. 2011). Clitellate embryos undergo a modified version of unequal spiral cleavage; individual blastomeres can be characterized reproducibly on the basis of size, position, the pattern of cell divisions by which they arise, and their subsequent fates (Weisblat and Huang 2001). Intriguingly, genomic analyses show extensive genomic rearrangements in Helobdella robusta relative to two other lophotrochozoan models (Cho et al. 2010; Simakov et al. 2013).
To begin elucidating mechanisms by which the germ line is specified in Helobdella embryos, we had previously identified and characterized the expression and function of a nanos homolog in H. robusta, finding that after broad early expression, it becomes restricted first to male presumptive PGCs and then later appears in female PGCs (Pilon and Weisblat 1997; Kang et al. 2002; Agee et al. 2006). Here, we have characterized piwi and vasa homologs in Helobdella: the H. robusta genome revealed two copies of each of these genes; orthologs were cloned and their expression characterized in H. austinensis (whose genes are designated with the prefix Hau-); as with Hro-nos, all copies of Hau-piwi and Hau-vasa are broadly expressed in early stages and gradually became restricted to PGCs as development unfolds, consistent with the GMP-PriSC model.
In contrast to our previous characterization of nanos, however, expression of piwi and vasa is first evident in the female germ line. Lineage tracing combined with in situ hybridization (ISH) revealed that as for the male germ line, female PGCs arise from segmental mesoderm. Thus, male and female germline fates separate only after 19 rounds of zygotic mitoses, much later than in several other invertebrate models. Combined with the capacity of some Helobdella species for self-fertilization, our results suggest a mechanism that would contribute to the extensive genomic rearrangements seen in H. robusta and to the rapid speciation of the genus Helobdella relative to other leeches (Oceguera-Figueroa et al. 2011).
Results
Sequence Retrieval and Phylogenetic Analyses
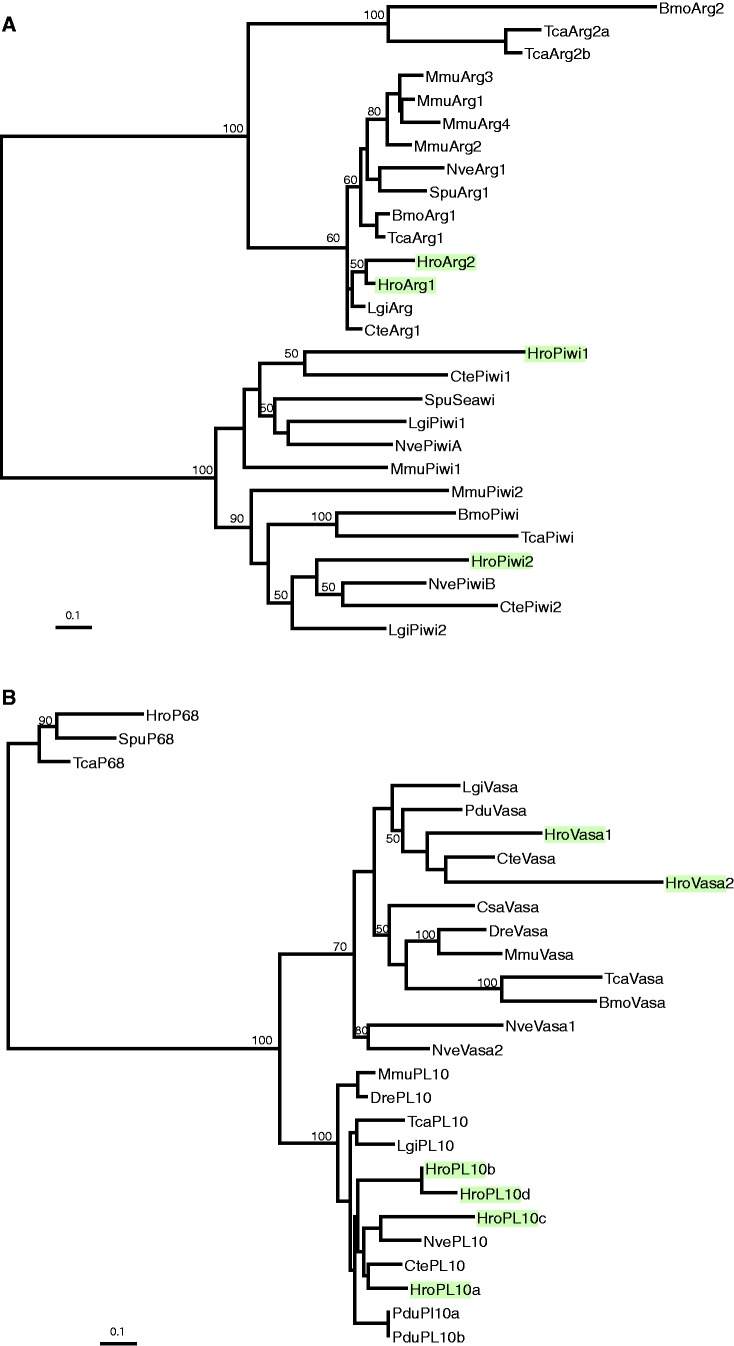
Three pairs of genes encoding homologs of Piwi, Vasa, and Argonaut were retrieved from the whole genome sequence of H. robusta, and four genes encoding PL10 homologs; we designate these genes Hro-piwi1, Hro-piwi2, Hro-vasa1, Hro-vasa2, Hro-ago1, Hro-ago2, and Hro-pl10a, Hro-pl10b, Hro-pl10c, and Hro-pl10d, respectively. Each set of these paralogs was distributed across different scaffolds with different flanking genes (supplementary table S1, Supplementary Material online); this suggests that the gene duplications were not of recent origin. To ask whether duplications of the piwi, vasa, and argonaut genes had occurred in an ancestor of annelids and mollusks, we also explored two other available lophotrochozoan genomes, the mollusk Lottia gigantea and the polychaete Capitella teleta. Similar to Helobdella, Lottia and Capitella, each have two piwi genes. In contrast to Helobdella, however, these other two lophotrochozoans each contain only one vasa, one Argonaut, and one pl10 gene, suggesting that these three genes may have undergone duplications in the sublineage leading from a last common annelid ancestor to leech.
Consistent with candidate identities, a maximum likelihood phylogenetic tree groups the piwi and argonaut genes in separate clades (fig. 1). The evolutionary dynamics of genes within these two clades differ, however. Although the arrangement of genes within the argonaut clade suggests that multiple, taxon-independent duplications of argonaut genes have occurred, the two clear clades separating metazoan piwi1 from piwi2 genes suggests a single, ancestral duplication of the piwi gene (fig. 1A). Similarly, the maximum likelihood phylogenetic tree encompassing the vasa and pl10 homologs groups these two sets of genes into separate clades as well. As in the case of the argonaut genes, several duplications appear to have occurred independently within both these gene clades (fig. 1B).
Fig. 1.
Molecular phylogenies for piwi and vasa genes. Maximum likelihood trees for (A) piwi and (B) vasa family genes in relation to the respective outgroups pl10 and argonaut. Helobdella robusta genes are indicated by shading. Numbers indicate bootstrap values equal to or greater than 50%. See text for details.
To study the expression of these piwi- and vasa-related leech genes, we cloned their orthologs from the sibling species H. austinensis, which is more easily maintained in laboratory culture. Coding sequences of the Hau-piwi and Hau-vasa genes were 97–98% similar at the nucleotide level when compared with their Hro-piwi and Hro-vasa orthologs, respectively, and 97–98% identical at the amino acid level.
Expression of Hau-piwi1 and Hau-vasa1 in the Adult Leech
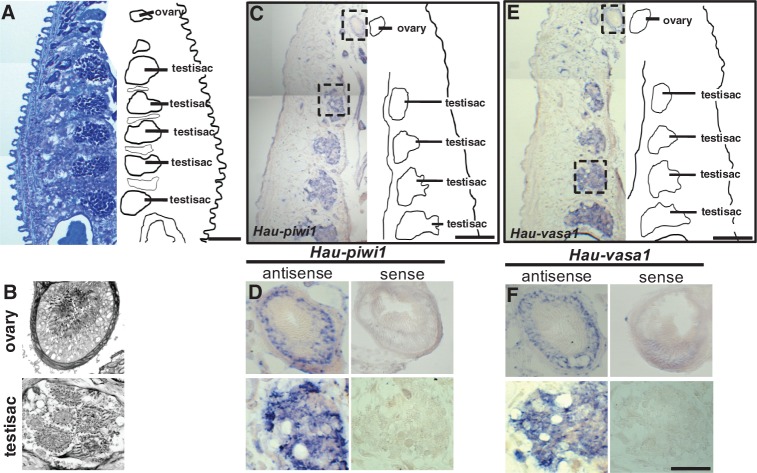
Leeches, including Helobdella, are simultaneous hermaphrodites, characteristic of clitellate annelids as a group. Leeches have one pair of ovaries (fig. 2A and B), associated with midbody segment 6 (designated as segment M6). The male gonopore lies in segment M5, but the number and distribution of the testis sacs varies among (and to a lesser extent within) species. Helobdella robusta and H. austinensis normally have six pairs of testis sacs, in segments M8–M13 (fig. 2A and B). To test the assumption that piwi and vasa are germ cell markers in the leech, we processed cryosectioned adult leeches by ISH for Hau-vasa1 and Hau-piwi1. Both genes were expressed specifically in the bilaterally paired ovaries and in the testis sacs (fig. 2C–F); sense probes gave no signal (fig. 2D and F). These results are consistent with a conserved function of piwi- and vasa-related genes in germ cell maintenance and establish their usefulness as germ cell markers in the leech.
Fig. 2.
Hau-piwi1 and Hau-vasa1 are expressed in gonads of the adult. Horizontal cryosections showing midbody segments of young adult Helobdella, stained with toluidine blue (A, B) or processed by ISH for Hau-piwi1 (C, D) or Hau-vasa1 (E, F). Anterior is up in all panels. For panels A, C, and E, the right side of each section is replaced by a line drawing indicating the reproductive (ovary, testisac) tissues. (A) Helobdella has one pair of ovaries, associated with segment M6, and six pairs of testisacs (five of which are visible here), associated with segments M8–M13. (B) Higher magnification views showing the arrangement of cells within a typical ovary and testisac. (C–F) Equivalent views of specimens processed by ISH show that Hau-piwi1 and Hau-vasa1 are expressed in both tissues. Boxes in (C) and (E) indicate areas shown at higher magnification in (D) and (F), respectively. Scale bar, 200 µm in (A), (C), (E); 50 µm in (B), (D), and (F).
Temporal and Spatial Patterns of Hau-piwi and Hau-vasa Expression in Development
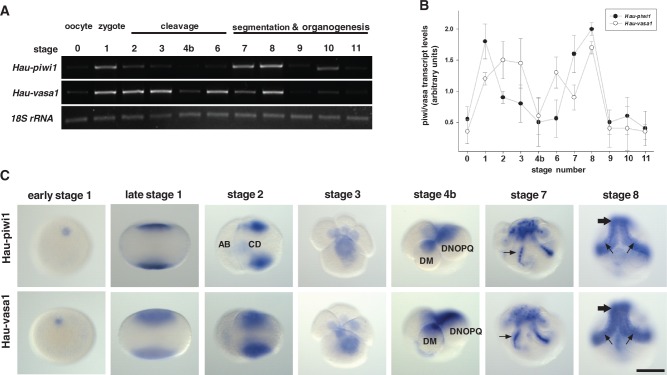
Leech development from deposition of the zygote (which is fertilized internally and then arrests at metaphase of meiosis I until deposition) to the juvenile has been divided into 11 stages (Fernández and Stent 1982). To obtain an overview of Hau-piwi1 and Hau-vasa1 expression, we performed reverse transcription polymerase chain reaction (RT-PCR) for each developmental stage as well as for the oocytes (fig. 3A and B). Both transcripts were detected at low levels in oocytes dissected from gravid individuals (stage 0) and in the final stages of development before the feeding juvenile (stages 9–11). Between these points, we observed two peaks of expression (fig. 3A and B). The first peak was associated with the zygote for Hau-piwi1 and with early cleavage in the case of Hau-vasa1. Both transcripts declined to low levels during third cleavage (stage 4b) and then peaked again during stages 7–8. This second peak correlated with the production of segmental founder cells (blast cells) in sequential, anteroposterior progression by the five bilateral pairs of lineage-restricted segmentation stem cells (teloblasts) that constitute a posterior growth zone in the embryos of clitellate annelids as described later.
Fig. 3.
Two peaks of Hau-piwi and Hau-vasa expression during early development. (A) Ethidium bromide-stained gel showing one of three replicate developmental RT-PCR experiments for Hau-piwi1 (30 cycles), Hau-vasa1 (30 cycles), and Hau-18S-rRNA (25 cycles). (B) Graph showing the average amount (±SD) of Hau-piwi1 (filled circles) and Hau-vasa (open circles) relative to Hau-18S-rRNA for the selected stages. (C) Representative embryos at selected stages that had been processed by ISH for Hro-piwi1 (top row) or Hro-vasa1 (bottom row): early stage 1 embryos (viewed obliquely) show transcript around the female pronucleus at the animal pole; stages late 1 and 2 (lateral views), 3 (animal pole view), and 4b (lateral view) show the association of both transcripts with teloplasm during cleavage (cells DNOPQ and DM are labeled); stage 7 (viewed obliqely with teloblasts and bandlets visible in dorsal posterior territory) show transcripts over nuclei (arrows), suggestive of active transcription; midstage 8 (viewed ventrally) shows broad accumulation of transcript throughout the germinal bands (small arrows) and germinal plate (large arrows). These data show an early peak of transcript accumulation in teloplasm, indicative of late maternal or very early zygotic transcription followed by decay to a minimum at the onset of fourth cleavage (stage 4b) and a rise in transcript levels during germinal band formation. Scale bar, 100 µm.
We have characterized the temporal and spatial expression of both duplicates of the piwi and vasa genes by ISH and found no significant differences between the expression of Hau-piwi1 and Hau-piwi2, or between Hau-vasa1 and Hau-vasa2. For brevity, only the results for Hau-piwi1 and Hau-vasa1 are presented below, but the following description applies to both piwi and both vasa paralogs.
Hau-piwi1 and Hau-vasa1 exhibited similar expression patterns at all developmental stages examined, from zygote to juvenile (stages 1–11). In the early zygote, both transcripts were confined to the region of the female pronucleus at the animal pole (fig. 3C). In the late zygote, the transcripts were associated with yolk-deficient domains of cytoplasm (teloplasm) at the animal and vegetal poles of the embryo (fig. 3C). Teloplasm is enriched in mRNAs and mitochondria segregated to the D quadrant by unequal cleavage (Fernández and Olea 1982; Fernández et al. 1987; Holton et al. 1989, 1994). It is required for D quadrant determination (Astrow et al. 1987). Hau-piwi1 and Hau-vasa1 mRNAs remained associated with teloplasm during cleavage but also appeared within the perinuclear cytoplasm in the macromeres.
Stage 4b begins with the division of macromere D′ (eight cell stage) to form blastomeres DM and DNOPQ, the precursors of mesodermal and ectodermal teloblasts, respectively. Both DM and DNOPQ inherit teloplasm. As in stage 3, both Hau-piwi1 and Hau-vasa1 transcripts at this stage were present in association with the teloplasm (of DM and DNOPQ) and the nucleoplasm of macromeres (fig. 3C).
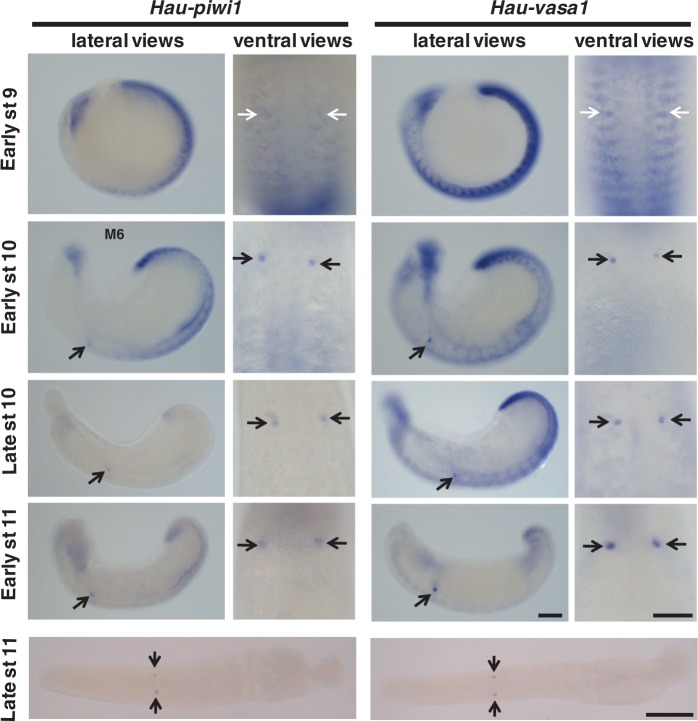
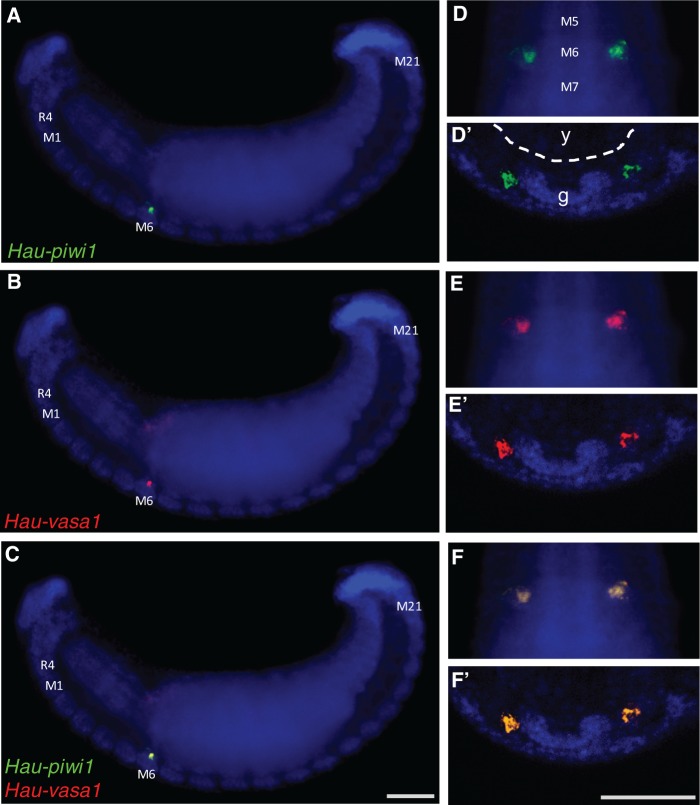
During blast cell (stages 6 and 7) and germinal band (stage 8) formation, transcripts were observed in all five teloblast lineages (mesodermal and ectodermal). The expression of both transcripts was strong and homogeneous throughout the bandlets during these stages (figs. 3C and 4). During stage 9, the levels of both vasa and piwi transcripts decreased in anteroposterior progression (fig. 4). The residual expression was not uniform and appeared as transverse stripes on either side of the midline in each segment. During stage 10, these stripes of expression largely disappeared as well, leaving a single bilateral pair of Hau-piwi1 and Hau-vasa1 positive spots in midbody segment 6 (segment M6), which we take to be the PGCs of the female germ line (figs. 4 and 5). Double-fluorescent ISH experiments confirmed that Hau-piwi and Hau-vasa were coexpressed in these cells (fig. 5). Staining in these two clusters of cells persisted through late stage 11, by which time the arms of the ovaries were visible in cross section (supplementary fig. S1, Supplementary Material online).
Fig. 4.
Restriction of Hau-piwi1 (left) and Hau-vasa1 (right) transcripts to female PGCs during later development. For each gene, the left column shows side views of whole embryos at stages 9–11, anterior to left, while the right column shows ventral views of 5–10 segments, anterior up. The bottom image in each column shows a ventral view of an intact juvenile (stage late11, anterior to left). Black arrows indicate the prospective female PGCs (segment M6) in selected images; white arrows indicate the corresponding position in early stage 9 embryos, at which time differential staining of the PGCs is not obvious. Scale bar, 100 µm.
Fig. 5.
Coexpression of Hau-piwi1 and Hau-vasa1 in female PGCs. Two color fluorescent in situs for Hau-piwi1 (green; A, D, D′) and Hau-vasa1 (red; B, E, E′) and merged views (C, F, F′). Nuclei were counterstained with DAPI (blue). (A, B, C) Lateral view, anterior to left, showing the entire early stage 11 embryo. Labels indicate rostral segment 4 (R4), and midbody segments 1, 6, and 21 (M1, M6, M21). (D, E, F) Ventral view, anterior up, showing midbody segments 5, 6, and 7 (M5, M6, M7). (D′, E′, F′) Transverse section, ventral down, through midbody segment 6; labels in (D') indicate yolk (y) within the developing midgut (dotted contour) and the segmental ganglion (g). Scales bars, 100 µm in (A–C); 50 µm in(D–F) and (D′–F′).
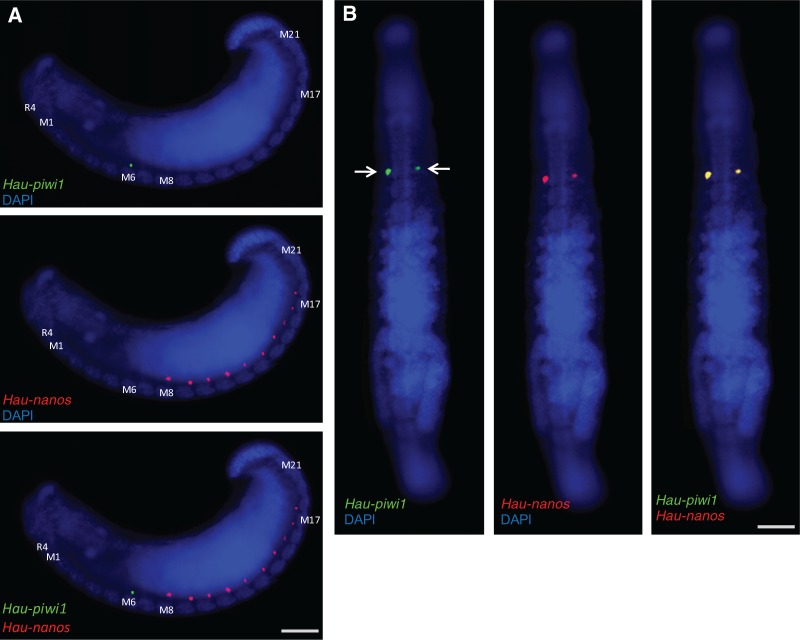
No expression of piwi or vasa genes was observed in the male primordia of segments M8–M18 at any of the developmental stages examined. Curiously, we have previously observed robust expression of another germline marker, namely a nanos-class gene, in presumptive male germ line primordia during stages 9 and 10, well before the nanos homolog was detected in the presumptive female germ line (Kang et al. 2002). From these two sets of observations, it would appear that in Helobdella, piwi and vasa are expressed preferentially in the developing female germline, while nanos is expressed preferentially in the male germline. We have confirmed this conclusion by double ISH for Hau-nanos and Hau-piwi. Consistent with our previous results, Hau-nos is expressed preferentially in the presumptive male germline precursors in early stage 11, with the difference that we observed only ten pairs of spots (segments M8–M17) in H. austinensis compared with 11 (segments M8–M18) in H. robusta (Kang et al. 2002; fig. 6A); by late stage 11, Hau-nos is coexpressed with Hau-piwi (fig. 6B) and Hau-vasa in the female germline precursors and has disappeared from the presumptive male germline precursors (fig. 6B).
Fig. 6.
Differential expression of Hau-nanos and Hau-piwi1 in the male and female PGCs, respectively. Two color fluorescent in situs for Hau-piwi1 (green) and Hau-nanos (red). (A) Lateral views of early stage 11 embryo; Hau-piwi1 is expressed in the female PGCs of segment M6 (top), Hau-nanos is expressed in male PGCs of segments M8–M17 (center) and there is no overlapping expression (bottom). (B) Ventral views of late stage 11 embryo; by this time, Hau-piwi1 (left) and Hau-nanos (center) are coexpressed in the female PGCs (right); Hau-nanos expression is not detected in the male germline at this stage. Scale bars, 100 µm in (A); 200 µm in (B).
Primorida Female Germline Cells Arise from Segmental Mesoderm in Helobdella
In leeches and other clitellate annelids, segmental mesoderm arises in a lineage-dependent manner by the interdigitation of stereotyped clones of segmental mesodermal (sm) blast cells, which arise from the stem cell divisions of M teloblasts (Weisblat and Shankland 1985; Goto et al. 1999). To test the hypothesis that the piwi-and-vasa-positive, female germline precursors in Helobdella arise from segmental mesoderm, as was found previously for the nanos-positive presumptive male germline precursors (Kang et al. 2002), we carried out fluorescent ISH for piwi and/or vasa on embryos in which M teloblasts had been injected with lineage tracers at various times.
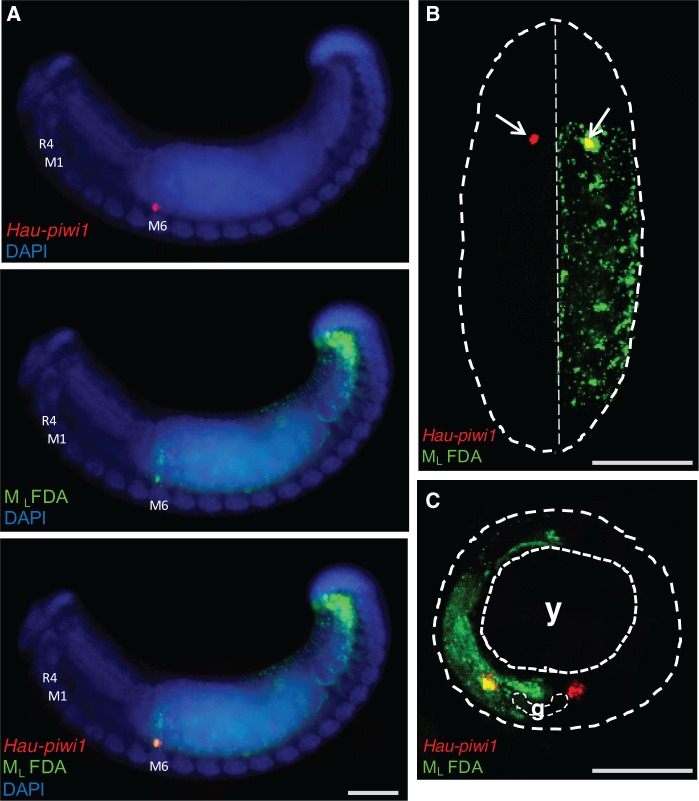
In one series of experiments, designed to test the hypothesis that the M teloblast lineage gives rise to the female germline, the newly formed left mesodermal teloblast (ML) was injected with FarRed dextran amine at stage 5, and the resultant embryos were examined by confocal microscopy at early stage 11, after processing by ISH with probes for piwi and vasa: the left cluster of piwi-and-vasa-positive cells in segment M6 was labeled with lineage tracer (supplementary fig. S2, Supplementary Material online), indicating that the female germline originates from the ipsilateral M lineage.
In a second series of experiments, designed to identify which M blast cell gives rise to the female germline, the left mesodermal teloblast (ML) was injected with fluorescein dextran amine (FDA) 15 cell cycles after its birth; by this point, the M teloblasts had each generated six early mesodermal blast cells (em1–em6), which contribute to nonsegmental tissues (Gline et al. 2011) and then nine sm blast cells (sm1–sm9), whose clones are located sequentially in the four rostral segments (R1–R4, associated with the anterior sucker) and the first five midbody segments (M1–M5). The embryos were examined by confocal microscopy at stage 11, after processing by ISH with probes for piwi: the clusters of piwi-expressing cells in segment M6 were labeled with lineage tracer on both sides in these embryos (fig. 7). On the left side, the piwi-positive cluster was in the first labeled segment, indicating that the female PGCs originate from blast cell sm10 in the M lineage.
Fig. 7.
Female PGCs originate from segmental mesodermal blast cell sm10. (A) Fluorescence micrographs of embryos in which the left mesodermal teloblast (ML) was injected with FDA (green) 15 cell cycles after its birth, i.e., after the M teloblasts had each generated six presegmental blast cells (em1–em6) and nine segmental blast cells (sm1–sm9). The embryos were examined by confocal microscopy at stage 11, after processing by ISH with probes for Hau-piwi1 (red). The piwi-expressing cell(s) in segment M6 were labeled with lineage tracer on the left side. (A) Lateral view showing left side, anterior to left. (B) Ventral view, anterior up showing roughly eight segments. Dotted contour outlines the visible portion of the embryo; on the left (apparent right) side, the piwi-positive cluster is in the first labeled segment, indicating that the female PGCs originate from blast cell sm10 in the M lineage. (C) Transverse section of a similar embryo. Coarse dotted contour outlines the embryo; finer contours outline the yolk (y) and the ventral ganglion (g), Scale bar, 100 µm in (A); 50 µm in (B); 30 µm in (C).
Discussion
Differential Expression of Conserved Germ Line Markers in Developing Male and Female PGCs
In the the work presented here, we have identified homologs of piwi and vasa, evolutionarily conserved germ line markers, in the genome of the leech H. robusta, and characterized their expression in embryos of the closely related H. austinensis. The piwi genes from both species group in distinct piwi1 and piwi2 clades with pairs of piwi genes from other species, consistent with a duplication of the piwi gene early in bilaterian evolution. By contrast, the two vasa genes in Helobdella appear to represent a more recent duplication not seen in two other available lophotrochozoan genome sequences, from the mollusc L. gigantea and the polychaete annelid C. teleta.
Leeches have discrete male and female reproductive organs, in contrast to the hybrid gonad (ovotestis) seen in most hermaphrodites; in Helobdella, a bilateral pair of ovaries is associated with midbody segment M6, and six pairs of testisacs lie in midbody segments M8–M13. In the adult, Hau-piwi and Hau-vasa are coexpressed in the gonads and testisacs as expected (Mochizuki et al. 2001; Extavour and Akam 2003; Oyama and Shimizu 2007; Rebscher et al. 2007; Dill and Seaver 2008; Kranz et al. 2010). Moreover, both piwi genes and both vasa genes were expressed in both the male and female germ line in the adult. Thus, we found no evidence to suggest that the duplicated genes had evolved sex-specific functions.
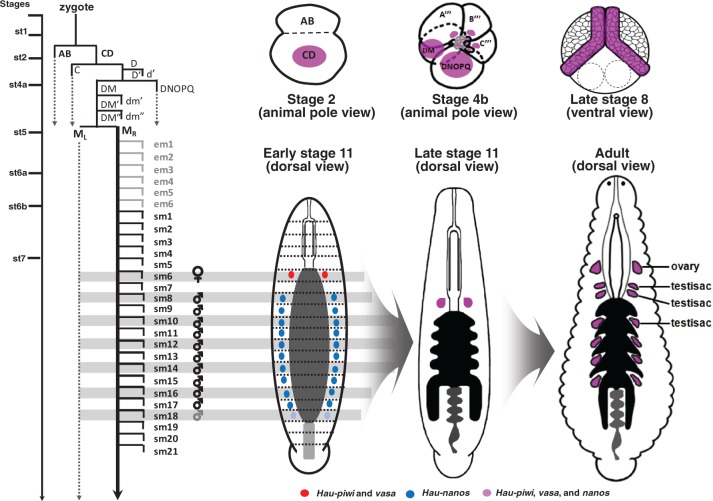
However, a striking result from our present work (summarized in fig. 8) is that, as levels of piwi and vasa decline during stage 9, their transcripts become restricted specifically to the female germ cells (segment M6), whereas nanos transcripts become restricted first to presumptive male germ line primordia during this same period (Kang et al. 2002). Thus, the differences do not arise simply from differences in the timing of the differentiation of the male and female organs but reflect differential expression of nanos in developing male and vasa and piwi in developing female germline. Intriguingly, these temporal differences in the expression of conserved germ line markers between the male and female germ cells correlate with greater developmental plasticity in male compared with the female germ line. First, as discussed previously, only six of the ten segments in which pairs of presumptive male germ line primordia express Hau-nos form definitive testisacs. Second, consistent with the predicted effects of social group size on sperm competition (Parker 1998), Helobdella exhibits postembryonic developmental adjustments in testisac volume, but not egg volume, depending on the number of animals in a mating group (Tan et al. 2004).
Fig. 8.
Diagrammatic summary of germ line marker expression and PGC segregation in Helobdella. Lineage diagram depicting cell divisions leading from the zygote to the segmental founder cells from which female and male PGCs arise, with line drawings showing the expression of conserved germ line markers at selected stages and in the adult leech (nanos, blue; coexpression of piwi and vasa red; coexpression of nanos, piwi and vasa, purple). Time line is approximate: from zygote to stage 7 takes about 54 h at 23 °C; teloblast cell cycles are 1–2 h. Bilateral M teloblasts arise at seventh cleavage from the division of DM″ (micromere 4d in standard nomenclature for spiralian embryos); each M teloblast undergoes six further rounds of stem cell division that produce nonsegmental early mesoderm blast cells (em1–em6) before producing its first segmental blast cell (sm1) (Gline et al. 2011); finally, six further rounds of mitosis occur before the precursor of the female germ line (sm6) are separated from the precursor of the male germ line (which arise from the M teloblast starting with sm8). As shown in figure 3C, coexpression of all three markers is prominent in teloplasm and teloblast precursors (but is also seen in other cells) during cleavage stages (e.g., stages 2 and 4b), and in the germinal bands and early germinal plate (stage 8). By early stage 11, expression of piwi and vasa has become restricted to the females PGCs (segment M6), while nanos is restricted to prospective male PGCs (segments M8–M17 in H. austinensis; and also segment M18 in H. robusta, indicated by paler shading). By late stage 11, expression of all three genes is restricted to the female PGCs. The point at which expression resumes in adult male PGCs remains to be determined. See text for further details. In the adult, ovaries are associated with segment M6 and definitive testisacs with segments M8–M13.
We are unaware of observations regarding the differential expression of germline markers in other hermaphrodites that form separate ovaries and testes as does leech. In the ovotestis of the nematode Caenorhabditis elegans, however, the piwi homolog prg-1 is required for spermatogenesis, and PRG-1 is preferentially expressed in the male portion of the ovotestis during the spermatogenesis phase (Wang and Reinke 2008). C. elegans has also undergone a marked expansion within the argonaut gene family, which contains more than 20 argonaut-family genes. Two of these genes, alg-3 and alg-4, are required semiredundantly for spermatogenesis and ALG-3 at least is preferentially expressed in the male portion of the ovotestis (Conine et al. 2010).
Delayed Segregation of Germ Line Precursors
Combined lineage tracing and situ hybridization shows that the presumptive female PGCs in Helobdella arise from the mesodermal lineage, specifically from the same M teloblast-derived blast cell that contributes most of the segmental mesoderm to midbody segment 6 (M6). We have previously shown that the presumptive male PGCs also arise from mesodermal blast cells (Kang et al. 2002). Thus, as summarized in figure 8, the male and female germ line fates in Helobdella fates separate from one another only after 19 rounds of zygotic mitoses (and from somatic lineages even later), much later than in other invertebrate models. For example, germline is set aside at third cleavage in the amphipod crustacean Parhyale hawaiensis (Gerberding et al. 2002), at fourth cleavage in the nematode C. elegans (Sulston et al. 1983), after nuclear division cycle eight in Drosophila melanogaster (Foe and Alberts 1983), at the eighth and ninth divisions in the polychaete annelid P. dumerilii (Rebscher et al 2007), and at the 11th division in the mollusc Crepidula fornicata (Lyons et al. 2012).
Leeches, together with earthworms and their relatives, comprise a monophyletic annelid subgroup called Clitellata (Erséus and Källersjö 2004; Struck et al. 2011). The PGCs in another clitellate annelid, the oligochaete Tubifex, also arise from the M teloblast lineages (Goto et al. 1999), but whether from segmental blast cells remains to be determined. Whether or not inductive signals are required to specify the PGCs during embryogenesis in these species remains to be determined, but restoration of germline following amputation in the oligochaete Enchytraeus (Sugio et al. 2008) must involve inductive signals of some sort. Together with the delayed segregation of germ line from somatic lineages in Helobdella, these results are consistent with the idea that some sort of epigenetic germ line specification was ancestral for this group (Extavour 2007).
Broad Early Expression of Germ Line Markers
The Helobdella piwi and vasa genes are broadly expressed during early development (summarized in fig. 8) and then are gradually restricted to presumptive germ line cells (see next section). We have previously observed a similar pattern of expression for Hro-nanos (Kang et al. 2002), and similar patterns of broad early expression becoming gradually restricted to the PGCs have been observed for one or more of these conserved germ line markers for other annelid genera, including Tubifex (Oyama and Shimizu 2007), Platynereis (Rebscher et al. 2007), Capitella (Dill and Seaver 2008; Giani et al. 2011), and Enchytraeus (Sugio et al. 2008). For Tubifex (like Helobdella, a clitellate annelid), it has been shown that restriction of vasa to the germ line represents primarily differential stabilization of preexisting transcripts (Oyama and Shimizu 2007); by contrast, our RT-PCR results show that in H. austinensis both piwi and vasa transcripts increase from the oocyte to the zygote or early cleavage stages, decline during mid-to-late cleavage, and then rise again during formation of the germinal bands and early germinal plate, indicative of a strong transcriptional component to their expression.
The early expression of piwi, vasa, and nanos homologs throughout the embryo is seen not only in annelids but also in mollusks (Swartz et al. 2008; Kranz et al. 2010), cnidarians (Extavour et al. 2005), flatworms (Shibata et al. 1999; Pfister et al. 2008), arthropods (Nakao et al. 2006; Mito et al. 2008; Ozhan-Kizil et al. 2009), and echinoderms (Juliano et al. 2010). In several cases, a functional requirement for these genes in early development distinct from germ line specification has also been demonstrated. In H. robusta for example, a partial antisense morpholino knockdown of nanos expression disrupts micromere positioning, germinal band elongation, and epiboly (Agee et al. 2006). It has been proposed that the broad early expression of piwi, vasa, and nanos homologs in both somatic and germline stem cells in flatworm and polychaete reflects a common origin for these cells during embryogenesis and that this may be the ancestral situation for metazoans (Rebscher et al. 2007). Elaboration of these ideas has led to the hypothesis of a broadly conserved germline multipotency program (GMP) involving extensive translational repression (Juliano et al. 2010), which functions not only in PGCs but also in an evolutionarily ancient type of PriSCs (Solana 2013), from which germ cells, somatic stem cells (neoblasts), and somatic tissues may arise. Our present results support these ideas.
Speculation on a Possible Developmental Basis for Acceleration of Genome Rearrangements in Helobdella Relative to Other Animals
Recent analyses of three lophotrochozoan genomes (Simakov et al. 2013) revealed a dramatic acceleration of gene rearrangements relative to protein evolution in the leech H. robusta that was not seen in the other species sequenced. Our present results suggest an interplay of developmental and reproductive strategies that might explain this.
Hermaphroditism is an ancestral trait for clitellate annelids. It is commonly held that clitellates are incapable of self-fertilization, but self-fertility has been observed for H. triserialis (Wedeen et al. 1990), H. robusta (Weisblat DA, unpublished observations), and H. papillornata (Tan et al. 2004), representing at least two distinct clades within the genus Helobdella (Siddall and Borda 2003; Bely and Weisblat 2006). Helobdella austinensis (Kutschera et al. 2013) is in the H. robusta clade (Bely and Weisblat 2006) but is not self-fertile (Levine M, Weisblat DA, unpublished data); given the apparent prevalence of self-fertility in the Helobdella genus, the self-infertility in H. austinensis may represent a secondary loss. Self-fertilization, an extreme form of inbreeding, provides a partial explanation for the accelerated genome rearrangements: otherwise lethal genetic translocations that move genes from one chromosome (linkage group) to another can be rescued by combining gametes that have the same translocation.
But this explanation alone is unsatisfactory. The nematode C. elegans is also a self-fertile hermaphrodite and has a much faster generation time (4 days for C. elegans vs. 9–10 weeks for H. robusta under laboratory conditions). Accordingly, its genome has diverged much more rapidly from the inferred ancestral protostome than has H. robusta by most criteria except that of genome rearrangements; traces of macrosynteny are still evident in the genome of C. elegans but not in that of Helobdella (Simakov et al. 2013).
Our analysis of germline formation in Helobdella suggests a plausible resolution to this paradox. Whereas Caenorhabditis segregates germline from somatic cells at the fourth zygotic mitosis (and generates male and female gametes by a somewhat indeterminate combination of proliferative and stem cell divisions thereafter; Kimble and Hirsch 1979), ipsilateral male and female PGC lineages in Helobdella are not segregated from each other until 19 rounds of zygotic mitosis have occurred (fig. 8). A priori, this extensively shared lineage increases the opportunity for genetic translocations to occur in cells that will contribute both male and female PGCs and which therefore might be rescued by self-fertilization. Conversely, self-fertilization selects against any homozygous lethal mutations that arise in a common precursor of male and female PGCs. Thus, the combination of reproductive strategy (self-fertile hermaphroditism) and developmental features (delayed segregation of male and female PGCs) seen in modern H. robusta suggests an explanation for how the lineage from which this species arose might have accumulated extensive gene rearrangements, while other aspects of its genome relatively remained stable.
Testing this explanation for the extensive rearrangements of the H. robusta genome relative to other animals is not possible at present, given the scant information regarding genome organization and reproductive strategies among related taxa. Similar challenges arise in interpreting results from other examples of self-fertilizing hermaphrodites. The killifish Kryptolebias marmoratus (Teleostei: Cyprinodontiformes: Rivulidae) and a sibling species K. ocellatus are the only vertebrate hermaphrodites for which self-fertilization has been well documented (Tatarenkov et al. 2009). Microsatellite analysis of four Kryptolebias species indicates that the capacity for self-fertilization in K. marmoratus and K. ocellatus has been maintained for several hundred thousand years or more (Tatarenkov et al. 2009); analysis of locally most common multilocus genotypes suggests that fertilization assurance rather than mulitlocus coadaptation is the driving force favoring self-fertility in this species (Avise and Tatarenkov 2012). Kryptolebias marmoratus shows chromosomal constancy over a large geographic range, but whether this represents recent, rapid spread or selection against the expected rearrangements cannot be determined (Sola et al. 1997).
In the present case, support for our explanation would be provided by finding that Helobdella is the only clitellate group to have evolved self-fertilization and is the only species in this group with such extensive genome rearrangements. Alternatively, if genomic rearrangments are a characteristic of clitellate annelids as a group, it could reflect earlier gains and losses of self-fertilization within the clitellate lineage, or simply more extensive inbreeding associated with fluctuating populations of hermaphroditic animals in ephemeral freshwater habitats.
Materials and Methods
Animals
A laboratory colony of the species newly described as H. austinensis (Kutschera et al. 2013) was raised and embryos were collected as described elsewhere (Gline et al. 2011).
Gene Identification, Cloning, and In Vitro Transcription
Putative PAZ and piwi domains (for Piwi and Argonaut) and DEAD/DEAH box helicase domains (for vasa and PL10) encoded by the annotated H. robusta genome (DOE, Joint Genome Institute) were identified by a Pfam search, using accession numbers PF02170, PF02171, and PF00270, respectively. As a further step, gene models of piwi/Ago and vasa/PL10 orthologs were used to perform BLASTP similarity search of the H. robusta genome. For each locus, the sets of gene models were assembled into a single predicted best-fit model for each gene using the SeqManII software (DNASTAR, Madison, WI).
Specific primers (piwi1 forward: ACCTCCATACTGGAGTATGATGAGGGAATA; piwi1 reverse: TAGTTTGTTGGCCAGAGTGTTACTAGGTGA; vasa1 forward: TTCCTGCTTCCTGTGTTGACGGGTATGTTG; vasa1 reverse: GCCACATCTACCAGTTCTACCAATCCTATG) were designed against the piwi1 and vasa1 gene models and amplified from cDNA prepared from the closely related H. austinensis. PCR products were gel extracted with a Qiagen Gel Extraction Kit and ligated into pGEM-T easy vector according to manusfacturer’s instructions. These clones were introduced into Escherichia coli DH105 competent cells by heat shock, plated onto solid media (15 g agarose per liter LB broth plus 50 µg/ml ampicillin) and grown overnight at 37 °C. Colonies were selected and grown overnight in LB broth with 1% ampicillin. Plasmids were isolated using the Qiagen Miniprep Kit following manufacturer’s instructions and sequenced commercially on an ABI platform. The MEGAscript (Ambion) kit was used to make digoxygenin (Dig-11-UTP) and/or biotin (Biotin-16-UTP) riboprobe according to manufacturer’s instructions.
Alignment and Phylogenetic Analyses
Piwi/Argonaut
A data set was built with orthologs of piwi and argonaut from representatives of the major metazoan clades with a total of 28 amino acid sequences (supplementary tables S1 and S2, Supplementary Material online). These sequences were prealigned using AMAP (Larkin et al. 2007; Schwartz and Pachter 2007) followed by manual editing in Se-Al v2.0a11 (http://tree.bio.ed.ac.uk/software/seal/, last accessed October 28, 2013). Gblocks were used to exclude poorly aligned amino acid positions under the least stringent parameters as implemented in the online version of the program (Castresana 2000). The resulting alignment comprised 512 amino acid positions (TREEBASE 14911 [http://purl.org/phylo/treebase/phylows/study/TB2:S14911, last accessed November 8, 2013]). The Whelan and Goldman (WAG) model of protein evolution (Whelan and Goldman 2001) with gamma distribution + invariant sites was implemented in all analyses as selected by ProtTest v1.4 as the model that best fits the data (Abascal et al. 2005). Maximum likelihood analyses were executed in RAxML-v7.0.4 (Stamatakis et al. 2005), for which support was obtained from 1,000 bootstrap replicates. Trees were viewed and edited in Dendroscope v2.2.2 (Huson et al. 2007).
Vasa/PL10
A matrix comprising 26 Vasa and PL10 orthologs representing the major metazoan clades was constructed. This data set (comprising 370 amino acid residues) was analyzed as for Piwi/Argonaut (see earlier).
Developmental RT-PCR
Total RNA samples were prepared from H. austinensis embryos at selected stages with RNA Wiz (Ambion) according to the manufacturer’s instructions, using 40 embryos for each stage. The total RNA obtained (3 µg) was reverse transcribed using a first-strand cDNA synthesis kit (BD Biosciences, Palo Alto, CA). The PCR mixture (50 µl) contained 10× Taq buffer, 0.3 U Taq polymerase (Perkin–Elmer), 2.5 μM of dNTPs, 5 pmol of each set of primers, and 50 ng of cDNA from the selected stages as template (fig. 2A). To amplify Hau-piwi1- and Hau-vasa1-specific fragments, a pair of PCR primers was designed as follows: Piwi1-F: 5′-CCCGATATTCCCAACATAAGGGTGAGAAA-3′, Piwi1-R: 5′-TATTCCCTCATCATACTCCAGTATGGAGGT-3′, Vasa1-F: 5′-TTCCTGCTTCCTGTGTT GACGGGTATGTTG-3′, Vasa1R: 5′-GCCACATCTACCAGTTCTACCATTCCTATG-3′. The PCR reactions were performed under the following cycling conditions: an initial denaturation at 94 °C for 5 min followed by 25–35 cycles of denaturation at 94 °C for 30 s, annealing at 60–65 °C (depending on the primer set) for 30 s, and elongation at 72 °C for 1 min, with a final elongation step at 72 °C for 10 min. A 10-µl aliquot of each PCR reaction was removed after 25 cycles, while the remaining material underwent five more cycles of amplification. Each primer set yielded a PCR product of 387 and 882 bp in length for Hau-piwi1 and Hau-vasa1, respectively. The 18S rRNA sequence was used as an internal standard (QuantumRNA, Ambion). The extent of amplification was chosen empirically to avoid saturation of the amplified bands. To quantify PCR products, each sample was run in a 1.5% agarose gel and stained with ethidium bromide. Band intensity was measured with an Alphaimager (Alpha Innotech Corp.) using the Alphaease (v3.3b) program.
Histology
To visualize differentiated testisacs and ovaries, young adult leeches were fixed in 4% formaldehyde in PBS overnight at 4 °C and mounted, sectioned, and dried as described by Cho et al. (2010). Cryosections were mounted on glass slides and stained with freshly filtered toluidine blue (1 mg/ml 2.5% aqueous sodium carbonate, pH 11.1) before placing the coverslip.
ISH Lineage Tracing and Cross Sectioning
All ISHs, double-fluorescent ISHs, lineage tracing, and cross sectioning were performed as previously described (Cho et al. 2010). Embryos were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) to visualize cell nuclei. Probe lengths were 1,848 bp for Hau-piwi1, and 876 bp for Hau-vasa1.
ISH on Cryosections for Adult Leech
Adult leeches were fixed at 4 °C overnight (4% paraformaldehyde in 1× PBS) and dehydrated in methanol series, equilibrated in 30% sucrose and mounted in OCT (Tissue-Tek) and stored frozen before sectioning on a cryostat (8-μm sections). Prepared sections were dried (50 °C, 30 min) and stored at −70 °C until further use. The specimens were incubated at 37 °C for 10 min in 20 μg/ml pronase E in PBT (PBS plus 0.1% Tween20, pH 7.4), then rinsed three times in PBT and incubated in 0.3% H2O2 in methanol at room temperature for 20 min. Sections were then postfixed for 20 min in 4% paraformaldehyde and finally rinsed twice in PBT. Prehybridization was carried out at 67 °C for 30 min in hybridization buffer (50% deionized formamide, 5× SSC, 1× Denhardt’s solution, 0.1% CHAPS, 100 μg/ml heparin, 0.1% Tween 20, 100 μg/ml tRNA). The prehybridization buffer was replaced with fresh hybridization buffer containing 2 ng/ml of the corresponding probe (Hau-piwi1 or Hau-vasa1) and specimens were incubated at 67 °C overnight. Washed specimens were incubated at room temperature for 1.5 h in 1% blocking reagents (Roche) in PBT, then incubated at 4 °C for 16 h with 1/1,000 Anti-DIG/AP antibody (Roche) in 1% blocking reagents. After incubation, the color reaction was carried out using nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indoyl-phosphate (Roche) by standard procedures. Stained specimens were dehydrated in ethanol, mounted in Fluoromount-G (SouthernBiotech), and examined by brightfield microscopy.
Supplementary Material
Supplementary figures S1 and S2 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1 GM 074619) and the National Science Foundation (I0S-0922792) to D.A.W. The authors thank Emina Begovic, Liam Holt, Ulrich Kutschera, Nik Putnam, Dan Rokhsar, and Oleg Simakov for helpful discussions, and members of the Weisblat lab for extensive discussions and assistance.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Agee SJ, Lyons DC, Weisblat DA. Maternal expression of a NANOS homolog is required for early development of the leech Helobdella robusta. Dev Biol. 2006;298:1–11. doi: 10.1016/j.ydbio.2006.04.473. [DOI] [PubMed] [Google Scholar]
- Astrow S, Holton B, Weisblat DA. Centrifugation redistributes factors determining cleavage patterns in leech embryos. Dev Biol. 1987;120:270–283. doi: 10.1016/0012-1606(87)90124-2. [DOI] [PubMed] [Google Scholar]
- Avise JC, Tatarenkov A. Allard's argument versus Baker's contention for the adaptive significance of selfing in a hermaphroditic fish. Proc Natl Acad Sci U S A. 2012;109:18862–18867. doi: 10.1073/pnas.1217202109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely AE, Weisblat DA. Lessons from leeches: a call for DNA barcoding in the lab. Evol Dev. 2006;8:491–501. doi: 10.1111/j.1525-142X.2006.00122.x. [DOI] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Valles Y, Giani VC, Jr, Seaver EC, Weisblat DA. Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol Biol Evol. 2010;27:1645–1658. doi: 10.1093/molbev/msq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Dill KK, Seaver EC. Vasa and nanos are coexpressed in somatic and germ line tissue from early embryonic cleavage stages through adulthood in the polychaete Capitella sp. I. Dev Genes Evol. 2008;218:453–463. doi: 10.1007/s00427-008-0236-x. [DOI] [PubMed] [Google Scholar]
- Erséus C, Källersjö M. 18S rDNA phylogeny of Clitellata (Annelida) Zool Scripta. 2004;33:187–196. [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CG. The molecular machinery of germ line specification. Mol Reprod Dev. 2010;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- Extavour CG. Evolution of the bilaterian germ line: lineage origin and modulation of specification mechanisms. Integr Comp Biol. 2007;47:770–785. doi: 10.1093/icb/icm027. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Pang K, Matus DQ, Martindale MQ. vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev. 2005;7:201–215. doi: 10.1111/j.1525-142X.2005.05023.x. [DOI] [PubMed] [Google Scholar]
- Fernández J, Olea N. Embryonic development of glossiphoniid leeches. In: Harrison FW, Cowden RR, editors. Developmental biology of freshwater invertebrates. New York: Alan R. Liss; 1982. pp. 317–361. [Google Scholar]
- Fernández J, Olea N, Matte C. Structure and development of the egg of the glossiphoniid leech Theromyzon rude: characterization of developmental stages and structure of the early uncleaved egg. Development. 1987;100:211–225. [Google Scholar]
- Fernández J, Stent GS. Embryonic development of the hirudinid leech Hirudo medicinalis: structure, development and segmentation of the germinal plate. J Embryol Exp Morphol. 1982;72:71–96. [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Stuydies of nuclear and cytoplasmic behavior during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Takamura K. Characterization of an ascidian DEAD-box gene, Ci-DEAD1: specific expression in the germ cells and its mRNA localization in the posterior-most blastomeres in early embryos. Dev Genes Evol. 2000;210:64–72. doi: 10.1007/s004270050012. [DOI] [PubMed] [Google Scholar]
- Gerberding M, Browne WE, Patel NH. Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development. 2002;129:5789–5801. doi: 10.1242/dev.00155. [DOI] [PubMed] [Google Scholar]
- Giani VC, Yamaguchi E, Michael MJ, Seaver EC. Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. EvoDevo. 2011;2:10. doi: 10.1186/2041-9139-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gline SE, Nakamoto A, Cho SJ, Chi C, Weisblat DA. Lineage analysis of micromere 4d, a super-phylotypic cell for Lophotrochozoa, in the leech Helobdella and the sludgeworm Tubifex. Dev Biol. 2011;353:120–133. doi: 10.1016/j.ydbio.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Kitamura K, Shimizu T. Cell lineage analysis of pattern formation in the Tubifex embryo. I. Segmentation in the mesoderm. Int J Dev Biol. 1999;43:317–327. [PubMed] [Google Scholar]
- Holton B, Astrow SH, Weisblat DA. Animal and vegetal teloplasms mix in the early embryo of the leech, Helobdella triserialis. Dev Biol. 1989;131:182–188. doi: 10.1016/s0012-1606(89)80049-1. [DOI] [PubMed] [Google Scholar]
- Holton B, Wedeen CJ, Astrow SH, Weisblat DA. Localization of polyadenylated RNAs during teloplasm formation and cleavage in leech embryos. Roux's Arch Dev Biol. 1994;204:46–53. doi: 10.1007/BF00189067. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Swartz SZ, Wessel GM. A conserved germline multipotency program. Development. 2010;137:4113–4126. doi: 10.1242/dev.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Pilon M, Weisblat DA. Maternal and zygotic expression of a nanos-class gene in the leech Helobdella robusta: primordial germ cells arise from segmental mesoderm. Dev Biol. 2002;245:28–41. doi: 10.1006/dbio.2002.0615. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsch D. The posteembryonic lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Komiya T, Itoh K, Ikenishi K, Furusawa M. Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994;162:354–363. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- Kranz AM, Tollenaere A, Norris BJ, Degnan BM, Degnan SM. Identifying the germline in an equally cleaving mollusc: vasa and nanos expression during embryonic and larval development of the vetigastropod Haliotis asinina. J Exp Zool B Mol Dev Evol. 2010;314:267–279. doi: 10.1002/jez.b.21336. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Langguth H, Kuo DH, Weisblat DA, Shankland M. Description of a new leech species from North America, Helobdella austinensis n. sp. (Hirudinea: Glossiphoniidae), with observations on its feeding behaviour. Zoosyst Evol. 2013;89:239–246. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lyons DC, Perry KJ, Lesoway MP, Henry JQ. Cleavage pattern and fate map of the mesnetoblast, 4d, in the gastropod Crepidula: a hallmark of spiralian development. Evodevo. 2012;3:21. doi: 10.1186/2041-9139-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito T, Nakamura T, Sarashina I, Chang CC, Ogawa S, Ohuchi H, Noji S. Dynamic expression patterns of vasa during embryogenesis in the cricket Gryllus bimaculatus. Dev Genes Evol. 2008;218:381–387. doi: 10.1007/s00427-008-0226-z. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- Nakao H, Hatakeyama M, Lee JM, Shimoda M, Kanda T. Expression pattern of Bombyx vasa-like (BmVLG) protein and its implications in germ cell development. Dev Genes Evol. 2006;216:94–99. doi: 10.1007/s00427-005-0033-8. [DOI] [PubMed] [Google Scholar]
- Oceguera-Figueroa A, Leon-Regagnon V, Siddall ME. DNA barcoding reveals Mexican diversity within the freshwater leech genus Helobdella (Annelida: Glossiphoniidae) Mitochondrial DNA. 2011;21(Suppl 1):24–29. doi: 10.3109/19401736.2010.527965. [DOI] [PubMed] [Google Scholar]
- Oyama A, Shimizu T. Transient occurrence of vasa-expressing cells in nongenital segments during embryonic development in the oligochaete annelid Tubifex tubifex. Dev Genes Evol. 2007;217:675–690. doi: 10.1007/s00427-007-0180-1. [DOI] [PubMed] [Google Scholar]
- Ozhan-Kizil G, Havemann J, Gerberding M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev Biol. 2009;327:230–239. doi: 10.1016/j.ydbio.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Parker GA. Sperm competition and the evolution of ejaculates: toward a theory base. In: Birkhead TR, Moller AP, editors. Sperm competition and sexual selection. San Diego (CA): Academic Press; 1998. pp. 3–54. [Google Scholar]
- Pfister D, De Mulder K, Hartenstein V, Kuales G, Borgonie G, Marx F, Morris J, Ladurner P. Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319:146–159. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- Pilon M, Weisblat DA. A nanos homolog in leech. Development. 1997;124:1771–1780. doi: 10.1242/dev.124.9.1771. [DOI] [PubMed] [Google Scholar]
- Rebscher N, Zelada-Gonzalez F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306:599–611. doi: 10.1016/j.ydbio.2007.03.521. [DOI] [PubMed] [Google Scholar]
- Schwartz AS, Pachter L. Multiple alignment by sequence annealing. Bioinformatics. 2007;23:e24–29. doi: 10.1093/bioinformatics/btl311. [DOI] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Borda E. Phylogeny and revision of the leech genus Helobdella (Glossiphoniidae) based on mitochondrial gene sequences and morphological data and a special consideration of the triserialis complex. Zool Scripta. 2003;32:23–33. [Google Scholar]
- Simakov O, Marletaz F, Cho SJ, et al. (25 co-authors) Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola L, Marzovillo M, Rossi AR, Gornung E, Bressanello S, Turner BJ. Cytogenetic analysis of a self-fertilizing fish, Rivulus marmoratus: remarkable chromosomal constancy over a vast geographic range. Genome. 1997;40:945–949. doi: 10.1139/g97-121. [DOI] [PubMed] [Google Scholar]
- Solana J. Closing the circle of germline and stem cells: the Primoridal Stem Cell hypothesis. EvoDevo. 2013;4:2. doi: 10.1186/2041-9139-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Struck TH, Paul C, Hill N, et al. (11 co-authors) Phylogenomic analyses unravel annelid evolution. Nature. 2011;471:95–98. doi: 10.1038/nature09864. [DOI] [PubMed] [Google Scholar]
- Sugio M, Takeuchi K, Kutsuna J, Tadokoro R, Takahashi Y, Yoshida-Noro C, Tochinai S. Exploration of embryonic origins of germline stem cells and neoblasts in Enchytraeus japonensis (Oligochaeta, Annelida) Gene Expr Patterns. 2008;8:227–236. doi: 10.1016/j.gep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Swartz SZ, Chan XY, Lambert DJ. Localization of Vasa mRNA during early cleavage of the snail Ilyanassa. Dev Genes Evol. 2008;218:107–113. doi: 10.1007/s00427-008-0203-6. [DOI] [PubMed] [Google Scholar]
- Tan GN, Govedich FR, Burd M. Social group size, potential sperm competition and reproductive investment in a hermaphroditic leech, Helobdella papillornata (Euhirudinea: Glossiphoniidae) J Evol Biol. 2004;17:574–580. doi: 10.1111/j.1420-9101.2004.00692.x. [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Lima SM, Taylor DS, Avise JC. Long-term retention of self-fertilization in a fish clade. Proc Natl Acad Sci U S A. 2009;106:14456–14459. doi: 10.1073/pnas.0907852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Wedeen CJ, Price DJ, Weisblat DA. Analysis of the life cycle, genome and homeo box genes of the leech, Helobdella triserialis. In: Stocum DL, Karr TL, editors. The cellular and molecular biology of pattern formation. New York: Oxford University Press; 1990. pp. 145–167. [Google Scholar]
- Weisblat DA, Huang FZ. An overview of glossiphoniid leech development. Can J Zool. 2001;79:218–232. [Google Scholar]
- Weisblat DA, Shankland M. Cell lineage and segmentation in the leech. Phil Trans R Soc B. 1985;312:39–56. doi: 10.1098/rstb.1985.0176. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.