Abstract
Although evolutionary studies of gene function often rely on RNA interference, the ideal approach would use reverse genetics to create null mutations for cross-species comparisons and forward genetics to identify novel genes in each species. We have used transcription activator-like effector nucleases (TALENs) to facilitate both approaches in Caenorhabditis nematodes. First, by combining golden gate cloning and TALEN technology, we can induce frameshifting mutations in any gene. Second, by combining this approach with bioinformatics we can predict and create the resources needed for forward genetic analysis in species like Caenorhabditis briggsae. Although developing genetic model organisms used to require years to isolate marker mutations, balancers, and tools, with TALENs, these reagents can now be produced in months. Furthermore, the analysis of nonsense mutants in related model organisms allows a directed approach for making these markers and tools. When used together, these methods could simplify the adaptation of other organisms for forward and reverse genetics.
Keywords: evolution, nematode, TALENs, model organism, C. briggsae
Introduction
With the advent of genome sequencing, there has been an explosion in the number of organisms being used for research into the evolution of gene regulation and function (e.g., Emlen et al. 2005) or that might be adapted for these studies in the near future (e.g., Kiontke et al. 2011; Kanzaki et al. 2012). However, the only method available for analyzing gene function in many species is RNA interference. Despite the simplicity of this approach, there can be significant variations in the response to RNAi between species (Nuez and Felix 2012), which complicates evolutionary comparisons.
The analysis of null mutations would provide a better method for comparing the functions of genes across species, but creating these mutations can be laborious, and maintaining them often requires balancers. In addition, forward genetic screens would allow the unbiased identification of new genes or genes with unexpected functions, which can be critical for evolutionary analysis (Guo et al. 2009). However, the ability to carry out these screens depends on genetic markers, balancing chromosomes and tools that took decades to develop for Drosophila melanogaster or Caenorhabditis elegans. For example, the related nematode C. briggsae has been studied for years, but random screens have only produced convenient sets of markers for half of its chromosomes. We show that transcription activator-like effector nuclease (TALEN) technology can overcome this problem.
New Approaches
We used TALENs to help develop the model organism C. briggsae for genetic and evolutionary studies. TALENs can be designed to target any DNA sequence (reviewed by Sun and Zhao 2013), and were recently tested in both C. elegans and C. briggsae (Wood et al. 2011; Lo et al. 2013). However, each TALEN requires 15–20 repeat units to bind a unique target, so we adapted golden gate shuffling to streamline the process of making custom TALENs for nematodes. Furthermore, new mutations can be difficult to study unless other genetic resources are available, so we used information from a set of C. elegans null mutations to identify and target genes needed to create markers, balancers, and tools for C. briggsae. This approach could be adapted to develop many other species as genetic model organisms.
Results and Discussion
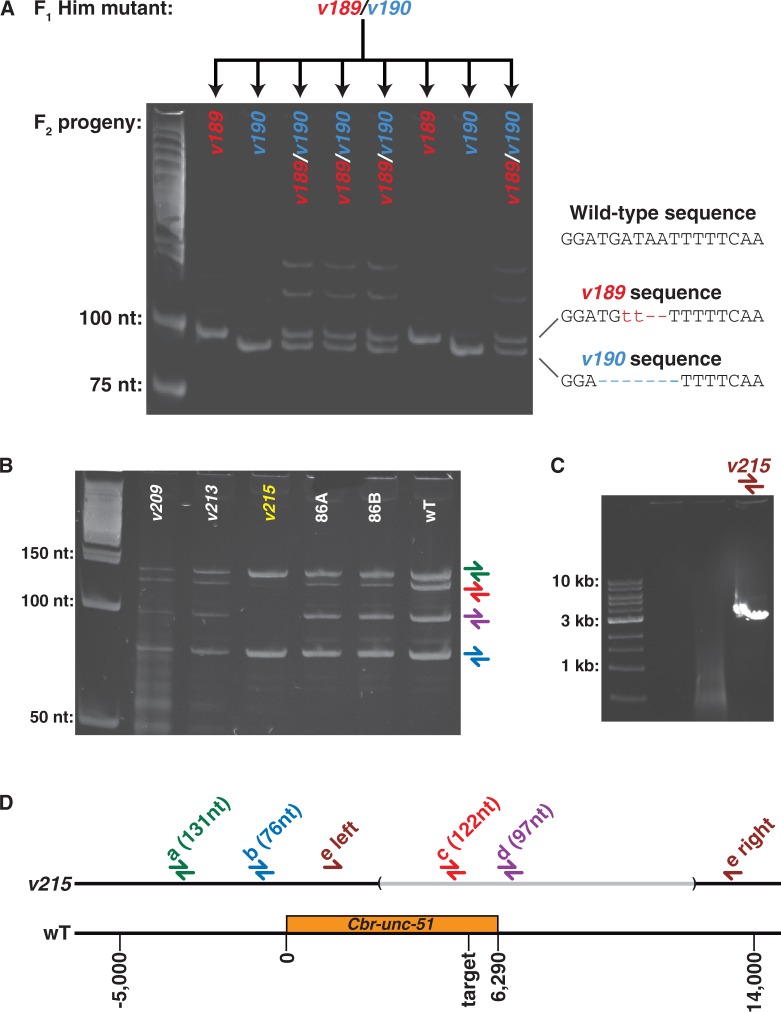
To simplify producing custom TALENs for C. briggsae, we adapted the golden gate shuffling method (Cermak et al. 2011) for use in nematodes (supplementary fig. S1, Supplementary Material online). Furthermore, to speed up the identification of new mutations, we screened for insertions or deletions by their altered size on 10% acylamide gels, eliminating the need for nuclease digestion (fig. 1A).
Fig. 1.
Isolation and characterization of TALEN-induced mutations in C. briggsae. (A) Many F1 animals were heterozygous for two distinct mutations. Following injection of TALENs targeting the Cbr-him-8 gene, we isolated an F1 animal with the Him phenotype (high incidence of male progeny). DNA from eight F2 self-progeny was amplified by the polymerase chain reaction (PCR) and separated on a 10% acrylamide gel, revealing the presence of two different alleles. The sequence of each lesion is shown at the right. (B) TALENs can produce large deletion mutations in nematodes. DNA from potential Cbr-unc-51 mutants was amplified using four sets of primer pairs (a–d). The size of each expected fragment is indicated in (D). Mutant Cbr-unc-51(v215) lacked DNA in the region of primer pairs c and d. (C) The primer pair e could amplify a single band of approximately 3 kb from v215 DNA, indicating that most of this region was deleted. (D) Map of the region, showing the extent of the 9423 bp v215 deletion, determined by sequencing the fragment shown in (C). Positions on the map are related to the start of Cbr-unc-51.
Using these approaches, we created hundreds of mutations in dozens of genes during the past 8 months (table 1, unpublished results). The efficiency of some TALENs was so high that many F1 animals were heterozygous for two new mutations: one induced in the maternal genome and a second in the paternal genome after fertilization (fig. 1A). Although most of the mutations were small insertions or deletions, we occasionally recovered large deletions (fig. 1B). Thus, TALENs can produce a variety of mutations in nematode genes, and the procedure is rapid and efficient.
Table 1.
Comparison of Caenorhabditis elegans and C. briggsae Mutations.
| Gene | LG | Protein | Cel Allele | Cel Phenotype | Cbr Allele | Cbr Phenotype | # Cbr Alleles |
|---|---|---|---|---|---|---|---|
| unc-40 | I | Netrin receptor (1,415 aa; Chan et al. 1996) | e1430 R157stop | Weak kinker Unc, slightly Dpy | v248fs (8 bp Δ) | Kinker Unc, slightly Dpy | 2 |
| dpy-5 | I | Procollagen (284 aa; Thacker et al. 2006) | e61 G203stop | Strong Dpy | v234fs (8 bp Δ/6 bp ins) | Strong Dpy | 28 |
| smg-5 | I | Novel (549 aa; Anders et al. 2003) | r860 Q17stop | NMD defective, pVul | v246fs (8 bp Δ) | NMD defective, pVul | 4 |
| him-8 | I | Zinc fingers (361; MacLeod et al. 1981) | tm611 deletion | High Incidence of males | v188fs (7 bp Δ) | High incidence of males | 9 |
| unc-54 | I | Myosin heavy chain (1,963 aa; MacLeod et al. 1981) | e1092 | Paralyzed | v139fs (11 bp Δ) | Paralyzed, lethal | 2 |
| Q1072stop | Unc | v138 (6 bp Δ) | Paralayzed Unc | ||||
| unc-34 | V | Enabled/VASP (468 aa; Yu et al. 2002) | gm104 W10stop | Coiler Unc | v255fs (11 bp Δ) | Coiler Unc | 7 |
| dpy-11 | V | Thioredoxin-like (246 aa; Ko and Chow 2002) | e207 R4stop | Dpy | v241 (6 bp Δ) | Dpy | 11 |
| unc-51 | V | Protein kinase (856 aa; Ogura et al. 1994) | ks38 Tc1 insertion | Paralyzed and Dpy | v204fs (5 bp Δ) | Paralyzed and Dpy | 14 |
| unc-1 | X | Stomatin-like (289 aa; Rajaram et al. 1998) | e719fs | Kinker Unc | v236fs (2 bp Δ) | Kinker Unc | 4 |
| dpy-8 | X | Collagen (452 aa; McMahon et al. 2003) | e130 unknown | Dpy | v262 (15 bp Δ) | Dpy | 8 |
| unc-7 | X | Innexin (522 aa; Krishnan et al. 1993) | e5 Q96stop | Kinker Unc | v272fs (11 bp Δ) | Kinker Unc | 3 |
Note.—If a gene produces more than one transcript, only the size of the largest product is listed. A representative null alleles or strong loss-of-function allele is shown for each gene. “fs” indicates a frameshift mutation, and “Δ” indicates a deletion. “Cel allele”—putative null alleles of each C. elegans gene except dpy-8, which lacked data. “Cbr allele”—reference alleles isolated in Caenorhabditis briggsae using TALENs. Most are null. “# Cbr alleles”—the number of alleles isolated in the TALEN screen. Some were not saved.
With this method in hand, we asked whether TALENs could be used to speed up the development of genetic model organisms, using C. briggsae as a test case. The first mutations in this species were isolated more than 60 years ago (Nigon and Dougherty 1950), but three of the six chromosomes still lacked sets of mapped marker genes to simplify genetic analysis. These three chromosomes (LG I, V, and X, www.briggsae.org, last accessed November 19, 2013) contain about 50 Mbp of DNA.
Fortunately, bioinformatic resources for C. briggsae are extensive. The genome sequence is nearly complete (Stein et al. 2003), and single nucleotide polymorphisms between the AF16 and HK104 strains have been used to create a rough linkage map (Hillier et al. 2007; Koboldt et al. 2010; Ross et al. 2011). These steps are critical for work in new species, but further progress depends on genetic resources. Unfortunately, forward screens have produced few visible markers, and new mutations take time to place on the genetic map. Reverse genetic approaches might solve this problem, but directed screens for large deletions have been slow and expensive (Hill et al. 2006). Thus, we applied TALEN technology.
Because TALENs usually produce small deletions, they often create null mutations. Thus, we tested two criteria for selecting potential marker genes in C. briggsae: 1) They must be orthologs of C. elegans genes with viable null mutations and 2) the C. elegans mutants must have visible phenotypes. For example, C. elegans unc-32(e189) is a widely used marker for LGIII, but unc-32 null alleles are lethal (Pujol et al. 2001), so we did not target its C. briggsae ortholog. Similarly, we avoided genes with wild-type null phenotypes, such as unc-93 (Greenwald and Horvitz 1980). After selecting potential markers, we targeted each gene with TALENs, to see whether these guidelines allowed an accurate prediction of phenotypes in C. briggsae (table 1). This effort represents one of the largest cross-species comparisons of animal gene function done using targeted knockouts.
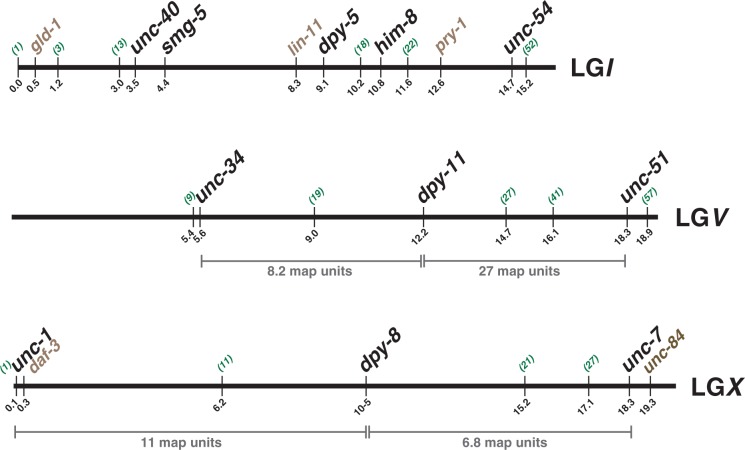
Using this approach, we rapidly isolated morphological mutants for the remaining C. briggsae chromosomes (fig. 2). The phenotypes of most mutants strongly resemble those of their C. elegans orthologs, including unc-40 and dpy-5 on LGI, unc-34 and unc-51 on LGV and unc-1, dpy-8, and unc-7 on X (table 1). However, the phenotypes of two genes were more severe—C. briggsae unc-54(v139fs) and dpy-11 null mutants were barely viable and could not be maintained, whereas the corresponding C. elegans mutants were much healthier. In these two cases, we also recovered partial loss-of-function mutations that were viable and easily scored. Overall, our approach predicted the null phenotypes for seven of nine genes and produced useful mutations for all nine. Moreover, each new mutation was already positioned in the C. briggsae genome, providing an instant link between the physical and genetic maps (fig. 2).
Fig. 2.
Engineering C. briggsae chromosomes for genetic analysis. Maps of the three C. briggsae chromosomes used in this project. Positions in megabases are shown below each line, as listed in Wormbase. The approximate positions in centimorgans of SNPs mapped by bulk segregant analysis are shown above each line in parentheses (Koboldt et al. 2010). Genes with mutations isolated in this paper are in large, bold letters. The locations of mutations in genes that affect vulval development, fertility, or dauer formation are shown in gray (Inoue et al. 2007; Seetharaman et al. 2010; Beadell et al. 2011; Sharanya et al. 2012).
To see if this method could be extended to other species, we chose C. sp. 11 (Kiontke et al. 2011), another hermaphroditic nematode. After identifying the C. sp. 11 unc-23 gene (supplementary fig. S2, Supplementary Material online), we used TALENs to induce v277 and v279, two frameshift mutations that are null alleles. The mutant animals have difficulty moving normally, often coil, and show progressively greater defects with age, because the head bends to one side. These phenotypes are very similar to those of C. elegans unc-23 mutants, which cause fragile muscle attachments (Plenefisch et al. 2000).
Although mutants with visible phenotypes can be used as balancers or to mark chromosomes in crosses, other genetic tools are also important. First, many TALEN mutants produce truncated proteins because of frameshifting mutations. If these transcripts are not eliminated by the nonsense-mediated decay (NMD) surveillance system (Weischenfeldt et al. 2005), they could be used to map functional domains in proteins. Thus, we used TALENs to knock out C. briggsae smg-5, which encodes a component of the NMD system (Anders et al. 2003). As predicted, this mutation restores partial activity to stop mutants in other genes (manuscript in preparation).
Second, hermaphroditic populations of nematodes reproduce by self-fertilization, but males are essential for crosses or phenotypic analysis. Thus, we created mutations in C. briggsae him-8, which controls X-chromosome pairing (Phillips and Dernburg 2006). Following self-fertilization, 35% of the Cbr-him-8 progeny were XO males and only 2% died as embryos (n = 498). These phenotypes are similar to those for C. elegans him-8 (Hodgkin et al. 1979).
Taken together, our results demonstrate a new use for TALEN or CRISPR (Damian and Porteus 2013) technologies—developing new species as genetic model organisms. This approach should be particularly valuable for species that are easily cultured in the laboratory and have closely related model organisms to guide the work, like members of Caenorhabditis, Pristionchus, Drosophila, or Tribolium. However, even distant model organisms might be helpful, because the proteins of C. elegans and C. briggsae are different from each other as those of humans and mice (Stein et al. 2003). Thus, information about Tribolium might aid in the development of more distant beetles as genetic model organisms, and zebrafish might be used for other fishes. However, there are some limits. For example, Xenopus laevis is polyploid, so it would be unsuitable for many genetic experiments. By contrast, TALENs work in the diploid frog X. tropicalis (Lei et al. 2013), so these techniques might facilitate its development as a model system.
For each new species, the critical steps are as follows: 1) Sequence the genomes of at least two inbred wild-type strains and identify SNPs; 2) develop a genetic map using a technique like advanced intercross recombinant inbred lines (Darvasi and Soller 1995); 3) use TALENs to create sets of genetic markers for each chromosome. Related species should provide a reliable guide, because most genes with nonsense mutations in C. elegans produced similar phenotypes when knocked out in C. briggsae; and 4) knock out additional genes to make genetic tools. Some tools would be determined by technical considerations, such as NMD mutations for analyzing truncations. Some would be determined by species, such as Him mutants for work with hermaphroditic nematodes, and others would be determined by the biological problem.
These steps are now rapid and inexpensive, usually less than $80 per gene for reagents. Opening up new species for sophisticated genetic analyses should revolutionize evolutionary developmental biology, because the use of null mutants provides a reliable way to compare gene function across species and nonbiased screens give a method for identifying evolutionary novelties (Guo et al. 2009).
Materials and Methods
Genetics
Caenorhabditis briggsae mutants were derived from the wild isolate AF16 (Fodor et al. 1983) and C. sp. 11 from JU1373 (Kiontke et al. 2011). Two-factor mapping was done as described by Brenner (1974). From dpy-8 unc-7/++ mothers, we saw 448 wild type, 19 Dpy, 22 Unc, and 131 Dpy Unc progeny. From dpy-11 unc-51/++, we observed 485 wild type, 96 Dpy, 105 Unc, and 72 Dpy Uncs. From unc-1 dpy-8/++, we observed 740 wild type, 63 Dpy, 62 Unc, and 333 Dpy Uncs, and from unc-34 dpy-11/++, we observed 1,008 wild type, 54 Dpy, 53 Unc, and 249 Dpy Uncs.
Procedures for Generating TALEN Knockout Mutants
First, pairs of custom TALENs were designed with TALE-NT 2.0 software (https://tale-nt.cac.cornell.edu/ [last accessed November 19, 2013]; Doyle et al. 2012), using a separation of 17 nt between binding sites. Each target sequence was 15–20 nt long and tested by Blast to avoid repetitive regions.
Second, each set of TALEN repeats was built using a golden gate assembly protocol (Cermak et al. 2011). Plasmids for the initial steps were purchased from Addgene (http://www.addgene.org/TALeffector/goldengateV2/, last accessed November 19, 2013). In the final step, the repeat sequences were cloned into the destination vector pRE189 (supplementary fig. S1, Supplementary Material online), rather than into one of the pTAL1-4 backbone vectors (Cermak et al. 2011). pRE189 combines sequences developed by Wood et al. (2011) with golden gate cloning sites and is optimized for use in Caenorhabditis nematodes.
Third, each plasmid was linearized by digestion with HindIII, treated with 100 µg/ml Proteinase K and 0.5% sodium dodecyl sulfate (SDS) for 30 min at 50 °C, and purified on a QIAquick column (Qiagen). We then synthesized mRNA using the SP6 mMessage Machine (Ambion) and purified it on MegaClear columns (Ambion). We precipitated each mRNA and dissolved it in water to give a final concentration of 4–6 µg/µl. Because pRE189 contains a transcribed polyA tail of 30 nt, we did not further polyadenylate the messages.
Fourth, the TALEN mRNAs were combined to produce a solution that was 2–3 µg/µl for each message. This solution was injected into the gonad of adult hermaphrodites (Wood et al. 2011), using methods developed by Evans et al. (1994). After injection, animals were soaked in recovery buffer (Evans 2006) and picked onto individual plates.
At 20 °C, the F1 progeny from a 6- to 32-h time window following the injection was singled to new plates, and F2 animals that carried mutations were identified by phenotype or by PCR analysis of the target site. Generally, we amplified fragments that were 60–100 nt long and separated them by size on 10% polyacrylamide gels (fig. 1A). Small insertions or deletions were obvious. More subtle changes were detected by the loss of a restriction site or the use of Cel1 nuclease (Wood et al. 2011).
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by National Institutes of Health grant GM085282 and National Science Foundation grant 1021128. We thank B. Meyer, T.-W. Lo, and T. Evans for reagents and advice.
References
- Anders KR, Grimson A, Anderson P. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell AV, Liu Q, Johnson DM, Haag ES. Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc Natl Acad Sci U S A. 2011;108:19672–19677. doi: 10.1073/pnas.1108068108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SS, Zheng H, Su MW, Wilk R, Killeen MT, Hedgecock EM, Culotti JG. UNC-40, a C. elegans homolog of DCC (deleted in colorectal cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 1996;87:187–195. doi: 10.1016/s0092-8674(00)81337-9. [DOI] [PubMed] [Google Scholar]
- Damian M, Porteus MH. A crisper look at genome editing: RNA-guided genome modification. Mol Ther. 2013;21:720–722. doi: 10.1038/mt.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlen DJ, Hunt J, Simmons LW. Evolution of sexual dimorphism and male dimorphism in the expression of beetle horns: phylogenetic evidence for modularity, evolutionary lability, and constraint. Am Nat. 2005;166(Suppl 4):S42–S68. doi: 10.1086/444599. [DOI] [PubMed] [Google Scholar]
- Evans TC. 2006. Transformation and microinjection. In: Wormbook editor. The C. elegans Research Community, Wormbook. doi/10.1895/wormbook.1.108.1. [Google Scholar]
- Evans TC, Crittenden SL, Kodoyianni V, Kimble J. Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Fodor A, Riddle DL, Nelson FK, Golden JW. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica. 1983;29:203–217. [Google Scholar]
- Greenwald IS, Horvitz HR. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lang S, Ellis RE. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr Biol. 2009;19:1853–1860. doi: 10.1016/j.cub.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 2006;10:531–538. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Ailion M, Poon S, Kim HK, Thomas JH, Sternberg PW. Genetic analysis of dauer formation in Caenorhabditis briggsae. Genetics. 2007;177:809–818. doi: 10.1534/genetics.107.078857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki N, Ragsdale EJ, Herrmann M, Mayer WE, Sommer RJ. Description of three Pristionchus species (Nematoda: Diplogastridae) from Japan that form a cryptic species complex with the model organism P. pacificus. Zoolog Sci. 2012;29:403–417. doi: 10.2108/zsj.29.403. [DOI] [PubMed] [Google Scholar]
- Kiontke KC, Felix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko FC, Chow KL. A novel thioredoxin-like protein encoded by the C. elegans dpy-11 gene is required for body and sensory organ morphogenesis. Development. 2002;129:1185–1194. doi: 10.1242/dev.129.5.1185. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Staisch J, Thillainathan B, Haines K, Baird SE, Chamberlin HM, Haag ES, Miller RD, Gupta BP. A toolkit for rapid gene mapping in the nematode Caenorhabditis briggsae. BMC Genomics. 2010;11:236. doi: 10.1186/1471-2164-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan SN, Frei E, Swain GP, Wyman RJ. Passover: a gene required for synaptic connectivity in the giant fiber system of Drosophila. Cell. 1993;73:967–977. doi: 10.1016/0092-8674(93)90274-t. [DOI] [PubMed] [Google Scholar]
- Lei Y, Guo X, Deng Y, Chen Y, Zhao H. Generation of gene disruptions by transcription activator-like effector nucleases (TALENs) in Xenopus tropicalis embryos. Cell Biosci. 2013;3:21. doi: 10.1186/2045-3701-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AR, Karn J, Brenner S. Molecular analysis of the unc-54 myosin heavy-chain gene of Caenorhabditis elegans. Nature. 1981;291:386–390. doi: 10.1038/291386a0. [DOI] [PubMed] [Google Scholar]
- McMahon L, Muriel JM, Roberts B, Quinn M, Johnstone IL. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol Biol Cell. 2003;14:1366–1378. doi: 10.1091/mbc.E02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigon V, Dougherty EC. A dwarf mutation in a nematode; a morphological mutant of Rhabditis briggsae, a free-living soil nematode. J Hered. 1950;41:103–109. doi: 10.1093/oxfordjournals.jhered.a106095. [DOI] [PubMed] [Google Scholar]
- Nuez I, Felix MA. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One. 2012;7:e29811. doi: 10.1371/journal.pone.0029811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Plenefisch JD, Zhu X, Hedgecock EM. Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development. 2000;127:1197–1207. doi: 10.1242/dev.127.6.1197. [DOI] [PubMed] [Google Scholar]
- Pujol N, Bonnerot C, Ewbank JJ, Kohara Y, Thierry-Mieg D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J Biol Chem. 2001;276:11913–11921. doi: 10.1074/jbc.M009451200. [DOI] [PubMed] [Google Scholar]
- Rajaram S, Sedensky MM, Morgan PG. Unc-1: a stomatin homologue controls sensitivity to volatile anesthetics in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:8761–8766. doi: 10.1073/pnas.95.15.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Koboldt DC, Staisch JE, Chamberlin HM, Gupta BP, Miller RD, Baird SE, Haag ES. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 2011;7:e1002174. doi: 10.1371/journal.pgen.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman A, Cumbo P, Bojanala N, Gupta BP. Conserved mechanism of Wnt signaling function in the specification of vulval precursor fates in C. elegans and C. briggsae. Dev Biol. 2010;346:128–139. doi: 10.1016/j.ydbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Sharanya D, Thillainathan B, Marri S, Bojanala N, Taylor J, Flibotte S, Moerman DG, Waterston RH, Gupta BP. Genetic control of vulval development in Caenorhabditis briggsae. G3. 2012;2:1625–1641. doi: 10.1534/g3.112.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, et al. (36 co-authors) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol Bioeng. 2013;110(7):1811–1821. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- Thacker C, Sheps JA, Rose AM. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell Mol Life Sci. 2006;63:1193–1204. doi: 10.1007/s00018-006-6012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Lykke-Andersen J, Porse B. Messenger RNA surveillance: neutralizing natural nonsense. Curr Biol. 2005;15:R559–R562. doi: 10.1016/j.cub.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, et al. (15 co-authors) Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Hao JC, Lim W, Tessier-Lavigne M, Bargmann CI. Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/enabled and a netrin-independent UNC-40/DCC function. Nat Neurosci. 2002;5:1147–1154. doi: 10.1038/nn956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.