Abstract
Membrane-associated GT-B glycosyltransferases (GTs) comprise a large family of enzymes that catalyze the transfer of a sugar moiety from nucleotide-sugar donors to a wide range of membrane-associated acceptor substrates, mostly in the form of lipids and proteins. As a consequence, they generate a significant and diverse amount of glycoconjugates in biological membranes, which are particularly important in cell–cell, cell–matrix and host–pathogen recognition events. Membrane-associated GT-B enzymes display two “Rossmann-fold” domains separated by a deep cleft that includes the catalytic center. They associate permanently or temporarily to the phospholipid bilayer by a combination of hydrophobic and electrostatic interactions. They have the remarkable property to access both hydrophobic and hydrophilic substrates that reside within chemically distinct environments catalyzing their enzymatic transformations in an efficient manner. Here, we discuss the considerable progress that has been made in recent years in understanding the molecular mechanism that governs substrate and membrane recognition, and the impact of the conformational transitions undergone by these GTs during the catalytic cycle.
Keywords: carbohydrate-modifying enzyme, glycosyltransferase, membrane protein, structural biology, X-ray crystallography
Introduction
Biological membranes play essential roles in nature. They are primarily used as physico-chemical barriers allowing cells to be functionally constituted and differentiated from the environment. Although biological membranes are stable structures, they are not static but highly dynamic in nature, allowing the selective transport of molecules across the cell, the modulation of the cellular response, as well as the occurrence of a diverse and important set of biochemical reactions. A typical membrane consists of a fluid phospholipid bilayer of 5–8 nm thickness in which a variety of proteins are embedded (Engelman 2005; McMahon and Gallop 2005). Phospholipids, the fundamental building blocks, are amphipathic molecules consisting of two hydrophobic fatty acid chains esterified at the sn-1 and sn-2 positions of glycerol, with a polar head group covalently linked to a phosphate moiety at the sn-3 position. They spontaneously form bilayers in aqueous solutions, in which the hydrophobic fatty acid tails are facing each other and are buried inside the membrane with the polar head groups exposed on both sides, in contact with water. Proteins, the other major component of biological membranes, associate permanently or temporarily with them, performing a diverse set of key functions in the cells. They account for ∼50% of the mass in plasma membranes, but their abundance can vary significantly according to the particular properties of the bilayer, reaching up to ∼75% in the inner membranes of mitochondria or chloroplasts. Glycans comprise 5–10% of the membrane weight mostly in the form of glycoconjugates, being covalently linked to both lipids and proteins. Importantly, all the three main constituents are represented by a substantial number of species, the amount and distribution of which also varies in space and time (Varki et al. 2009). Thus, membrane diversity provides their identity to cells, cellular compartments and vesicles, strongly influencing the development and maintenance of a myriad of forms of life.
Glycoconjugates are prominent components of biological membranes primarily localized on the surface of eukaryotic and prokaryotic cells (Spiro 2002; Trombetta and Parodi 2003; Helenius and Aebi 2004; Kowarik et al. 2006; Hattrup and Gendler 2008; Varki et al. 2009; Nothaft and Szymanski 2010). They are critical not only in the maintenance of the structural integrity of cell membranes but also in the modulation of molecular recognition events including cell-signaling, cell–cell and cell–pathogens interactions (Raetz and Whitfield 2002; Bos et al. 2007; Paulic and Bertozzi 2008; Weidenmaier and Peschel 2008). Most of the enzymes encoded in eukaryotic and prokaryotic genomes that are responsible for the biosynthesis and modification of these membrane-associated glycoconjugates are glycosyltransferases (GTs; Lairson et al. 2008). GTs catalyze the stereo- and regiospecific transfer of a sugar moiety from nucleotide-sugar or lipid-phospho-sugar donors to a wide range of acceptor substrates including lipids and proteins. In this review, we focus our attention on the membrane-associated GT-B family of GTs. We present the remarkable progress made in recent years on the understanding of the molecular and structural bases of catalysis and substrate and membrane association of this family of enzymes.
GTs: An overview
Catalytic mechanisms in GTs
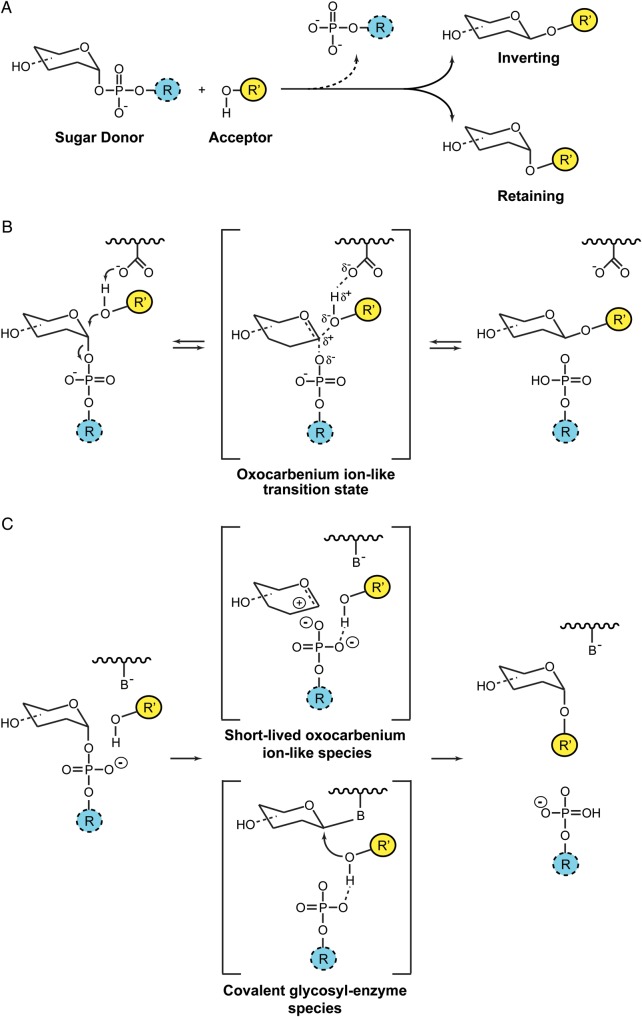
Glycosyl transfer can proceed with either “inversion” or “retention” of the anomeric configuration with respect to the reaction substrates and products (Lairson et al. 2008; Figure 1A). “Inverting” GTs seem to follow a single-displacement mechanism with an oxocarbenium ion-like transition state and an asynchronous SN2 mechanism, analogous to that observed for inverting glycosyl hydrolases (Figure 1B; Kakuda et al. 2004; Kozmon and Tvaroska 2006; Davies et al. 2012). In contrast, the catalytic mechanism for “retaining” enzymes remains a matter of strong debate. By analogy with glycosyl hydrolases, a double-displacement mechanism via the formation of a covalent glycosyl-enzyme intermediate was first suggested. Such a mechanism would involve an enzymatic nucleophile positioned within the active site on the β-face of the donor substrate in close proximity to the anomeric reaction center. In support of this model, the chemical rescue of a mutant form of a retaining α3-galactosyltransferase (α3GalT) by sodium azide has been reported (Monegal and Planas 2006). The product of this chemical rescue is the inverted version of the sugar azide, which is consistent with the first step in a double-displacement mechanism. Furthermore, covalent intermediates were directly detected for the human blood group synthesizing α-(1 → 3)-N-acetylgalactosaminyltransferase (GTA) and α-(1 → 3)-galactosyltransferase (GTB) mutants by mass spectrometry (Soya et al. 2011). However, in the absence of other clear experimental evidence of a viable covalent intermediate, an alternative mechanism known as the SNi “internal return” has been proposed (Persson et al. 2001; Gibson et al. 2002; Lairson et al. 2008). In this model, the departure of the leaving group and the nucleophilic attack occur on the same face of the sugar, involving a single-step mechanism through the formation of an oxocarbenium-like transition state with asynchronous C1-O acceptor glycoside bond formation and C1-O phosphate bond breakdown (Vetting et al. 2008; Frantom et al. 2010; Gomez et al. 2012). This concept has further evolved to a SNi-like mechanism claiming for the existence of a short-lived oxocarbenium ion intermediate allowing the nuclear rearrangement for proper acceptor attack (Ardévol and Rovira 2011; Lee, Lee et al. 2011; Lee, Hong et al. 2011). The current scenario distinguishes two groups of retaining GTs depending on the presence or absence of a putative nucleophile in the active site (Figure 1C; Rojas-Cervellera et al. 2013). In the case that a putative nucleophile is absent (e.g., OstA), the electrostatic potential of the active site is such that it can stabilize the oxocarbenium ion-like intermediate for a very short period. However, this time is long enough for the active site to reorganize, allowing the oxocarbenium-ion species and the acceptor to move one toward the other. In contrast, retaining GTs in which a putative nucleophile is present (e.g., α3GalT, GTA and GTB), the oxocarbenium ion-like transition state is stabilized by the formation of a covalent bond with a carboxylate residue. Therefore, both models could be considered as a variation of a common mechanism, in which a two-step reaction takes place with the formation of an oxocarbenium ion-like transition state. The intermediate might be stabilized via the formation of an oxocarbenium ion or a covalent glycosyl-enzyme depending on the particular structure of the active site (Rojas-Cervellera et al. 2013). Whether or not “retaining” GTs could proceed via different catalytic mechanisms is a notion that clearly needs further experimental support.
Fig. 1.
Catalytic mechanisms in GTs. (A) GTs catalyze the transfer of sugars with either “inversion” or “retention” of the anomeric configuration with respect to the sugar donor substrates. (B) Inverting GTs utilize a direct-displacement SN2-like reaction mechanism involving a single oxocarbenium ion-like transition state. (C) Current mechanisms for enzymatic glycosyl transfer with retention of configuration proposed in the literature: The front-face mechanism and the double-displacement mechanism (Rojas-Cervellera et al. 2013).
Structural folds in GTs
The GT-A and GT-B folds
Two major structural folds have been described for the nucleotide sugar-dependent enzymes among the first 40 GT sequence-based families (CAZy, Carbohydrate-Active enZymes data base; see www.cazy.org) for which three-dimensional (3D) structures have been reported. These topologies are variations of “nucleotide binding domains” and have been defined as GT-A and GT-B (Vrielink et al. 1994; Charnock and Davies 1999; Coutinho et al. 2003; Figure 2A and B). The “nucleotide binding” or “Rossmann-fold” domain contains two sets of β-α-β-α-β units (654123 topology), together forming a single parallel sheet flanked by α-helices. A long loop, or cross-over, frequently between strands 3 and 4 creates a natural cavity that participates in the binding of the nucleotide ring. The “Rossmann-fold” domain was first described for the lactate dehydrogenase from Squalus acanthus and found in a wide range of nucleotide-binding proteins including the uridine diphosphategalactose-4-epimerase and dehydropterin oxidoreductase (Rossmann et al. 1974; Lesk 1995).
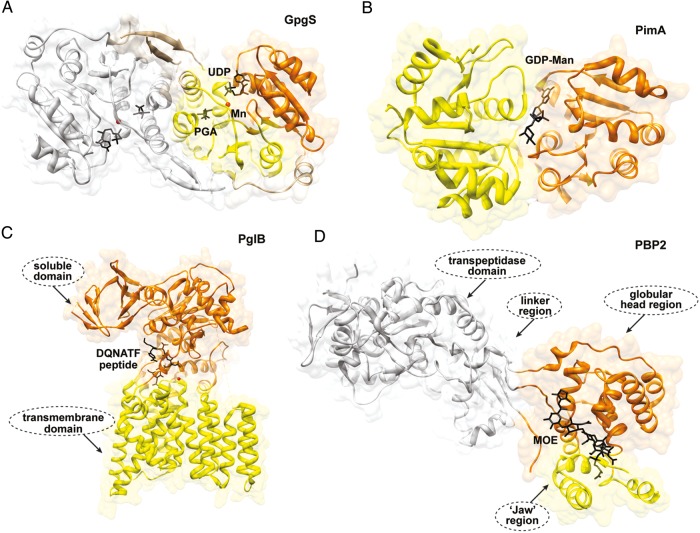
Fig. 2.
Structural folds in GTs. (A) The overall architecture of the GT-A fold as observed in the dimeric glucosyl 3-phosphoglycerate synthase from M. tuberculosis. The N- and C-terminal domains are shown in orange and yellow, respectively. The second monomer is shown in gray (Urresti et al. 2012). (B) The GT-B fold as visualized in the phosphatidyl-myo-inositol mannosyltransferase PimA from Mycobacterium smegmatis. The N- and C-terminal domains are shown in yellow and orange, respectively (Guerin et al. 2007). (C) GT-C fold members are predicted to have 8–13 transmembrane α-helices with the active site located at the interface between the transmembrane (yellow) and soluble (orange) domains as observed in PglB from Campylobacter lari (Lizak et al. 2011). (D) The peptidoglycan PBP2 from Staphylococcus aureus adopts a fold distinct from those of other GT classes (Lovering et al. 2007).
The GT-A fold consists of two tightly associated “Rossmann-fold” domains, the sizes of which may vary, leading to the formation of a continuous β-sheet. The N-terminal domain participates in the recognition of the nucleotide sugar donor, whereas the C-terminal domain interacts mainly with the acceptor substrate. Most GT-A enzymes exhibit an Asp-Xaa-Asp (also known as DXD) signature in which one or both carboxylate groups coordinate a divalent cation in order to stabilize the pyrophosphate group of the donor substrate (Hu and Walker 2002; Lairson et al. 2008). Specific loops, adjacent to the active site, often adopt different conformations and appear to play a crucial role during substrate binding and catalysis (Ramakrishnan et al. 2004; Urresti et al. 2012). The GT-B fold was first described for the 351-amino acid DNA-modifying β-glucosyltransferase from family GT63, an inverting GT from bacteriophage T4, and was found to be structurally related to the catalytic core of glycogen phosphorylase (Barford and Johnson 1989; Vrielink et al. 1994; Artymiuk et al. 1995; Wrabl and Grishin 2001). During the last 5 years, the crystal structures of a significant number of GT-B enzymes have been reported (Table I). The GT-B fold displays two “Rossmann-fold” domains separated by a deep cleft that includes the catalytic center. Therefore, an important interdomain movement has been predicted or demonstrated in some members of this superfamily during substrate binding and catalysis including MurG (Hu et al. 2003), glycogen synthase (Buschiazzo et al. 2004; Sheng et al. 2009; Baskaran et al. 2010), PimA (Guerin et al. 2007; Guerin et al. 2009) and MshA (Vetting et al. 2008). It is generally accepted that in GT-B enzymes, the nucleotide-sugar donors mainly bind to the C-terminal domain of the protein, whereas the N-terminal domain is involved in acceptor substrate recognition. Since acceptors exhibit a marked diversity of chemical structures compared with nucleotide-sugar donors, the N-terminal domains reflect this variability by showing different rearrangements of secondary structural elements (Breton et al. 2006). In contrast to GT-A enzymes, structural and kinetic evidence indicate that divalent cations are not essential for enzymatic activity (Abdian et al. 2000; Lairson et al. 2008). However, the rates are accelerated by certain cations for reasons that are not yet understood (Hu and Walker 2002). On the basis of primary sequence homology analysis, it has been suggested that a glycogen phosphorylase/glycosyltransferase family motif is present in many GT-B enzymes (Abdian et al. 2000; Wrabl and Grishin 2001). However, GT-B enzymes do not seem to share any strictly conserved residues (Hu and Walker 2002). Both sequential ordered as well as random kinetic reactions have been described/proposed for enzymes belonging to the GT-B family. In the absence of membranes, MurG utilizes a compulsory ordered Bi-Bi mechanism in which the sugar donor UDP-N-acetylglucosaminyltransferase (GlcNAc) binds first, prior to the binding of the lipid acceptor (Chen et al. 2002). The glycosylated acceptor is then released, followed by the UDP group. Similarly, structural and kinetics evidence suggest that MshA proceeds by an ordered mechanism with UDP-GlcNAc binding first and 1-l-inositol-1-phosphate binding second (Vetting et al. 2008). In contrast, OleD, a GT that glycosylates oleandomycin, was shown to utilize an ordered mechanism wherein the acceptor substrate binds first (Quiros et al. 2000). Finally, structural and kinetic evidence suggest that the β-glucosyltransferase (BGT) from the T4 bacteriophage could bind the sugar donor UDP-Glc or the acceptor DNA in any order (Larivière and Moréra 2004). Interestingly, many of the structurally uncharacterized nucleotide sugar-dependent enzyme families are also predicted to adopt one of these 2-folds, suggesting that they might have evolved from a small number of progenitor sequences. In that sense, primitive archaea exhibit two GT families, GT2 (GT-A fold) and GT4 (GT-B fold) (Coutinho et al. 2003).
Table I.
GT-B GTs of known 3D structure
| GT-B Family | Catalytic mechanism | Membrane associated | Nonmembrane associated |
|---|---|---|---|
| GT1 | Inverting |
Alg13/Alg14 (PMa,
apo, Wang et al.
2008), Ugt2b7 (BTb, apo, Miley et al. 2007) |
CalG1/CalG2/CalG4, CalG3, EryCIII, GtfA, GtfB, GtfD, OGT/NGT, OleD/OleI, SpnG, Ufgt, UGT71G1, UGT78G1, UGT85H2, UrdGT2 |
| GT3 | Retaining | – | Gsy2 |
| GT4 | Retaining | AviGT4 (apo and UDP, Martinez-Fleites et al. 2006), CGT (MT, apo, Lee, Lee et al. 2011; Lee, Hong et al. 2011), PimA (PM, GDP and GDP-Man, Guerin et al. 2007), PimB’ (PM, GDP and GDP-Man, Batt et al. 2010), SUS1 (PM, UDP-Glc, UDP/Fru, Zheng et al. 2011), WaaG (MTc, UDP and UDP-2F-Glc, Martinez-Fleites et al. 2006), WsaF (PM, apo, dTDP and dTDP-Rha, Steiner et al. 2010), WbaZ (MT, apo, Liu et al. 1993) | BshA, MshA, NY2A_B736L, SpsA, TreT |
| GT5 | Retaining | – | AtGlgA, EcGlgA, PaGlgA, OsGBSSI, HvSSI |
| GT9 | Inverting |
WaaC (MT, apo, ADP and
ADP-2F-Hep, Grizot et al. 2006),
WaaF (MT, apo, pdb code
1PSW), Vpar_0760 (MT, apo, to be published) |
– |
| GT10 | Inverting | FucT (PM, apo, GDP and GDP-Fuc, Sun et al. 2007) | – |
| GT20 | Retaining | – | OtsA |
| GT23 | Inverting | FUT8 (BTb, apo, Ihara et al. 2007) | NodZ |
| GT28 | Inverting | MurG (MT, apo and UDP-GlcNAc, Hu et al. 2003; Brown et al. 2013) | – |
| GT30 | Inverting | WaaA (MT, apo and CMP, Schmidt et al. 2012) | – |
| GT35 | Retaining | – | GP (liver/muscle), Gph1, MalP, SP |
| GT41 | Inverting | – | HsOGT, XcOGT, HMW1C |
| GT52 | Inverting | NST (BT, apo, CMP, CDP and CMP-3F-Neu5Ac, Lin et al. 2011) | – |
| GT63 | Inverting | – | BGT |
| GT65 | Inverting | – | PoFUT1 |
| GT68 | Inverting | – | PoFUT2 |
| GT70 | Inverting | GumK (MT, apo, UDP, Barreras et al. 2008) | – |
| GT72 | Retaining | – | AGT |
| GT80 | Inverting |
PmST1 (BT, CMP and
CMP-3F-Neu5Ac, CMP-3F-Neu5Ac/Lac, CMP/Lac, Ni et al. 2007; Kim et al. 2008), ST (MT, CMP, Iwatani et al. 2009; Tsukamoto et al. 2007), Pst6-224 (MT, CMP, Kakuta et al. 2008) |
– |
aPM, peripheral GT; bBT, bitopic GT; cMT, monotopic GT; dn.d., not determined. TagF was classified as a GT2 enzyme due to the presence of both GT-A and GT-B folds in the GTs.
Alg13, UDP-GlcNAc:Dol-PP-GlcNAc N-acetylglucosaminyltransferase; Ugt2b7, UDP-GlcA:β-glucuronosyltransferase 2B7; CalG1, calicheamicin GT 1; CalG2, calicheamicin GT 2; CalG3, calicheamicin GT 3; CalG4, calicheamicin GT 4; EryCIII, TDP-desosamine:α-mycarosyl erythronolide B desosaminyltransferase; GtfA, dTDP-β-L-4-epi-epivancosamine:epivancosaminyltransferase; GtfB, TDP/UDP-glucose:aglycosyl-vancomycin glucosyltransferase; GtfD, UDP-β-l-4-epi-vancosamine:vancomycin-pseudoaglycone vancosaminyltransferase; OGT/NGT, UDP-Glc:sinapoyl-alcohol-, 2,5-DHBA-, 3,4-DHBA-glucosyltransferase; OleD, oleandomycin GT; OleI, oleandomycin GT; SpnG, TDP-β-l-Rha:spynosin 9-O-α-l-rhamnosyltransferase; Ufgt, UDP-Glc: Anthocyanidin 3-O-glucosyltransferase; UGT71G1, UDP-Glc:flavonoid β-glucosyltransferase; UGT78G1, UDP-glucose:flavonoid β–glucosyltransferase; UGT85H2, UDP-glucose:(iso)flavonoid β–glucosyltransferase; UrdGT2, urdamycin A GT II; Gsy2, glycogen synthase; CGT, cholesterol α-glucosyltransferase; PimA, GDP-Man:phosphatidylinositol mannosyltransferase; PimB’, GDP-Man: Phosphatidylinositolmannose mannosyltransferase; WaaG, UDP-Glc:l-glycero-d-manno-heptose II -1,3-glucosyltransferase I; WbaZ, putative mannosyltransferase; WsaF, TDP-β-L-Rha; S-Layer glycoprotein β-1,2-rhamnosyltransferase; WaaF, heptosyltransferase II; BshA, UDP-GlcNAc: l-malate α-N-acetylglucosaminyltransferase; MshA, UDP-GlcNAc:inositol-P N-acetylglucosaminyltransferase; TreT, trehalose synthase; SpsA, sucrose phosphate synthase; AviGT4, eurekanate-attachment enzyme; SUS1, sucrose synthase; NY2A_B736L, putative mannosyltransferase; AtGlgA, glycogen synthase; EcGlgA, glycogen synthase; PaGlgA, glycogen synthase; OsGBSSI, rice granule bound starch synthase; HvSSI, barley starch synthase I; WaaF, LPS heptosyltransferase II; WaaC, heptosyltransferase I; Vpar_0760, putative heptosyltransferase; FucT α-1,3-fucosyltransferase; OtsA, α,α-trehalose-phosphate synthase; Fut8, N-acetyl-d-glucosaminide -1,6-l-fucosyltransferase; NodZ, α-1,6-l-fucosyltransferase; MurG, UDP-GlcNAc: N-acetylmuramyl-(pentapeptide)-PP-C55 N-acetylglucosaminyltransferase; WaaA, CMP-β-KDO: α-3-deoxy-d-manno-2-octulosonic-acid (KDO) transferase; GP, glycogen phosphorylase; Gph1, glycogen phosphorylase; MalP, maltodextrin phosphorylase; SP, starch phosphorylase; NST, CMP-Neu: (LOS) β-galactosamide α-2,3-sialyltransferase; BGT, UDP-Glc: DNA β-glucosyltransferase; GumK, UDP-GlcA: (xanthan) α-Man-(1,3)-β-Glc-(1,4)- α-Glc-PP-polyisoprenyl β-1,2-glucuronosyltransferase; AGT, UDP-Glc: DNA α-glucosyltransferase; PmST1, CMP-NeuAc: α-2,3/2,6-sialyltransferase 1; ST, CMP-NeuAc: a-/β-galactoside α-2,3-sialyltransferase; Pst6-224, CMP-NeuAc: β-galactoside α-2,6-sialyltransferase (for further details, see www.cazy.org).
The GT-C fold
A third GT fold named GT-C was predicted on the basis of iterative sequence alignments (Oriol et al. 2002; Liu and Mushegian 2003). This important group of GTs comprises integral membrane proteins with 8–13 transmembrane helices, with dependency for lipid phosphate-activated donor sugar substrates (Berg et al. 2007; Lairson et al. 2008). The first reported crystal structure of a full-length member of the GT-C superfamily was that of PglB from Campylobacter lari, a close homolog of the eukaryotic oligosaccharyltransferase STT3 catalytic subunit (Lizak et al. 2011). The structure reveals that the transmembrane domain is indispensible both for peptide binding and for catalysis (Figure 2C). More recently, the crystal structure of MraY from Aquifex aeolicus has been solved at 3.3 Å resolution (Chung et al. 2013). MraY, which belongs to the polyprenylphosphate N-acetylhexosamine-1-phosphate transferase superfamily, is an integral membrane enzyme that catalyzes an essential step of bacterial cell wall biosynthesis: The transfer of the peptido-glycan precursor phospho-MurNAc-pentapeptide to the lipid carrier undecaprenyl phosphate.
Both inverting and retaining enzymes were found in GT-A and GT-B folds suggesting that there is no correlation between the overall fold of GTs and their catalytic mechanism. To date, all enzymes predicted to adopt the GT-C fold belong to the inverting GT family (Lairson et al. 2008). Interestingly, a considerable number of sequence families are not predicted to adopt the GT-A, GT-B or GT-C folds. In this respect, the crystal structure of a peptidoglycan GT from family GT51, an enzyme that utilizes a lipid-phospho-sugar as donor, revealed that this protein adopts a bacteriophage-lysozyme-like fold (Figure 2D; Lovering et al. 2007; Yuan et al. 2007). The structural characterization of these “orphan” families will certainly contribute to the understanding of the variety of folds that nature selectively uses to catalyze glycosyl transfer.
Membrane-associated GT-B GTs
Membrane proteins: An overview
Membrane proteins can be classified as either “peripheral” or “integral” according to their degree of association to the lipid bilayer (Luckey 2008). Peripheral membrane proteins attach temporarily to one face of the lipid bilayer or other membrane proteins. They interact weakly with the membrane mainly by noncovalent interactions including electrostatic and hydrogen bonds. As a consequence, peripheral membrane proteins can be removed by using relatively gentle treatments such as high ionic strength and alkaline buffers, leaving the lipid bilayer intact (Luckey 2008). In contrast, integral membrane proteins hold tightly and permanently to the membrane and can only be removed by treatment with detergents and organic solvents that disrupt the lipid bilayer (White and Wimley 1999; Andersen and Koeppe 2007). In addition, and based on their mode of insertion, integral membrane proteins can be classified into monotopic, bitopic and polytopic proteins (Blobel 1980). Monotopic proteins associate firmly to only one side of the lipid bilayer, whereas bitopic and polytopic proteins span the membrane by one or several transmembrane segments, respectively (Elofsson and von Heijne 2007). To date, two major structural architectures have been described for polytopic transmembrane proteins, α-helical and β-barrels. Most α-helical proteins are present in the membrane of archaea, the inner membrane of bacteria and the plasma and internal membranes in eukaryotes, whereas the β-barrels are found in the outer membrane of Gram-negative bacteria and in the outer membrane of chloroplast and mitochondria. Recently, several crystal structures for GT-B membrane-associated GTs have been solved, including peripheral and integral proteins (Table I).
Membrane association: Working at the membrane–water interface
Membrane-associated GT-B enzymes represent a specialized group of GTs with the remarkable ability to recognize both the hydrophilic water-soluble nucleotide mono- or diphospho-sugar donor and acceptor substrates mostly in the form of hydrophobic lipids and membrane-associated proteins (Figure 3; Forneris and Mattevi 2008). What are the structural requirements for a GT-B enzyme to be associated to the lipid bilayer? What is the impact of the membrane composition and structure on enzymatic activity? How is the active site of a membrane-associated GT-B GT made accessible to hydrophobic and hydrophilic substrates? Here, we focus on the two best-studied families of membrane-associated GT-B GTs: The monotopic and peripheral enzymes.
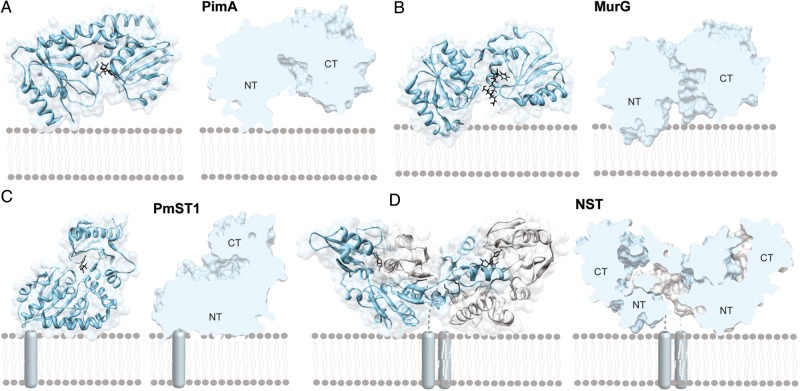
Fig. 3.
Membrane-associated GT-B GTs. (A) A GT-B peripheral membrane GT as illustrated by the phosphatidyl-myo-inositol mannosyltransferase PimA (PM, GT4; Guerin et al. 2007; Guerin et al. 2009). (B) A GT-B monotopic GT as illustrated by the UDP-N-acetylglucosamine–N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase MurG (MT, GT28; Hu et al. 2003) (C) A GT-B bitopic GT as illustrated by the multifunctional sialyltransferase PmST1 from Pasteurella multocida (Ni et al. 2006; Ni et al. 2007). (D) The GT-B dimeric monotopic α-2,3-sialyltransferase NST from Neisseria meningitidis (Gilbert et al. 1997; Lin et al. 2011).
Monotopic GT-B GTs
The 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii (AlMGS) represents a good model reflecting our current knowledge of the molecular mechanism of membrane association for the monotopic GT-B enzymes. Acholeplasma laidlawii is a cell wall-less prokaryote that controls the surface charge density and curvature properties of its membrane through the action of two cytosolic side membrane-associated glucosyltransferases (Dahlqvist et al. 1995; Andersson et al. 1996; Lindblom et al. 1986; Lindblom et al. 1993; Andrés et al. 2012). Specifically, the AlMGS synthase and the diglucosyl-diacylglycerol synthase (AlDGS) synthesize, respectively, the two major nonbilayer-prone and bilayer-forming glucolipids, GlcDAG and GlcGlcDAG, by consecutively adding two glucose residues to a diacylglycerol acceptor backbone at the membrane interface, respectively (Karlsson et al. 1997; Vikstrom et al. 1999). The molar ratios of these glucolipids and acylated counterparts vary markedly in response to a variety of stimuli, including acyl chain types and ionic environment (Wieslander et al. 1986). AlMGS lack a hydrophobic trans-membrane segment, but detergents are needed for its solubilization (Li et al. 2003). Interestingly, the N-terminal domain of AlMGS displays a calculated pI value of 9.7 and hence is positively charged at physiological pH. In contrast, the C-terminal domain of the enzyme is dominated by the presence of acidic residues (Berg et al. 2001; Edman et al. 2003). A close inspection of its amino acid sequence revealed that the N-terminal domain of the enzyme contains a mixture of hydrophobic and basic residues, mainly in the form of arginine and lysine, in an amphiphatic α-helix (Lind et al. 2007). The amphipathic character of this α-helix was structurally verified by a combination of solution nuclear magnetic resonance (NMR) and circular dichroism, and proved to interact with both zwitterionic and anionic phospholipids by surface plasmon resonance (SPR; Lind et al. 2007). Very recently, a preferential binding of AlMGS to anionic lipids including phosphatidylglycerol (PG) and cardiolipin (CL) was observed in vivo (Ariöz et al. 2013).
Membrane lipid surfaces are negatively charged essentially due to the presence of anionic lipids (McLaughlin 1989; McLaughlin and Murray 2005). The binding of AlMGS to lipid bilayers containing anionic lipids (e.g., phosphatidylglycerol) was measured by SPR and found to be essentially irreversible with an overall dissociation constant (KD) of 10−10–10−12 M (Vikstrom et al. 1999; Li et al. 2003). Moreover, the thickness of vesicular membrane fractions (lipid bilayer + interfacial water phase) containing AlMGS was determined to be ∼4.30 nm by small angle X-ray diffraction (Eriksson et al. 2009; Ge 2011). Since the thickness of the inner membrane of Escherichia coli is about 3.75 nm and the size of AlMGS is roughly 4 × 5 nm, these observations are suggestive of the deep insertion of AlMGS in the membrane, as also indicated by molecular dynamic simulation of other monotopic membrane proteins (Mitra et al. 2004; Balali-Mood et al. 2009). Thus, experimental data clearly point to a permanent association to the bilayer surface, without any apparent shuttling of AlMGS between the cytoplasm and the membrane, typically observed for peripheral membrane-associated proteins (Li et al. 2003; Lind et al. 2007). Interestingly, anionic phospholipids seem to affect the binding and also the enzymatic activity of AlMGS. None of the uncharged GlcGlcDAG, the neutral-charged 1,2-dioleoyl-phosphatidylcholine and positively charged sphingosine lipids could replace 1,2-dioleoyl-phosphatidylglycerol (DOPG) as an activator for the purified AlMGS (Karlsson et al. 1997). In contrast, the anionic phospholipid 1,2-dioleoyl-phosphatidylserine could fully substitute for DOPG, indicating that the anionic-charged groups of the activator lipids hold the activating property (Karlsson et al. 1994; Dahlqvist et al. 1995).
In plants, photosynthetic membranes are mainly constituted by two galactolipids, the nonbilayer-prone GalDAG and the bilayer-prone GalGalDAG (Dormann and Benning 2002; Kelly and Dörmann 2004; Guskov et al. 2009; Kobayashi et al. 2013; Ge 2011). The relative amounts of these two glycolipids affect the membrane curvature as well as the lateral stress profiles of the membrane, thus likely influencing the functions of other membrane-associated proteins or protein complexes (Moellering and Benning 2011). Arabidopsis thaliana synthesizes GalDAG through the action of a monogalactosyldiacylglycerol synthase, in the form of three GT-B isoforms namely AtMGD1, AtMGD2 and AtMGD3 (Benning and Ohta 2005). The three isoforms transfer a galactose residue from UDP-Gal to the 3-position of sn-1,2-diacylglycerol (DAG) and differ in their substrate specificity and subcellular localization. Two GT-B enzymes were found to catalyze the biosynthesis of GalGalDAG, AtDGD1 and AtDGD2 (Ge 2011). AtDGD2, a monotopic enzyme displaying similar interface-binding structural elements as AlMGS, is apparently able to modulate the lipid environment by adjusting the biosynthetic activity of its product GalGalDAG (Ge et al. 2011). Nonbilayer-prone lipids such as GlcDAG, rac-1,2-dioleoylgly-cerol and dioleoyl-sn-glycero-3-phosphoethanolamine decreased the enzymatic activity of AtDGD2, suggesting that increasing curvature stress with these additives has a negative effect on GalGalDAG synthesis. In contrast, the anionic lipids PG, phosphatidic acid and phosphatidylserine displayed strong stimulatory effects on AtDGD2 activity; interestingly, CL did not even though it is structurally related to PG, suggesting the existence of PG-specific-binding site(s) in the protein. Three segments of AtDGD2 containing conserved tryptophan residues were demonstrated to be crucial not only for enzymatic activity but also for membrane interaction (Ge et al. 2011; Szpryngiel et al. 2011). It is worth noting that the enrichment of tryptophan residues in the lipid bilayer interface regions is well established for many membrane-associated proteins (Kozlov 2010). Interestingly, two of the tryptophan-containing segments (IV and V) of AtDGD2 are localized in the N-terminal domain, whereas another is in the C-terminal domain (VI), with all segments likely interacting with stimulatory anionic phospholipids and determining the orientation of AtDGD2 at the membrane interface. Altogether the experimental data allowed Wieslander et al. to propose a regulatory model for the anchoring of AtDGD2 at the membrane interface (Figure 4). According to this model, the N-terminal segments IV and V may serve as permanent anchor points for the N-terminal domain at the lipid interface while segment V on the C-terminal domain interacts with the membrane interface in a more flexible way. The positively charged protein clusters located at the lipid interface are thought to play a determinant role in regulating the interaction of the whole enzyme with the membrane, leading to the different catalytic efficiencies (Ge 2011; Ge et al. 2011).
Fig. 4.
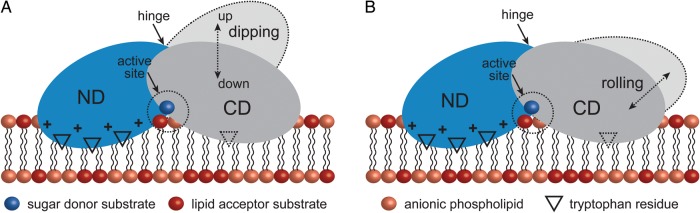
A model for membrane association of monotopic GT-B GTs. Wieslander and colleagues proposed two modes of reorientation for AtDGD2 upon stimulation by anionic lipids: An up-down displacement (“dipping”) of the C-terminal domain (A) and a “rolling” transfer of the catalytic region into the interface (B). Both models contemplate the access of the soluble UDP-Gal donor substrate to the active site.
The relevance between electrostatic forces, both coulombic attraction/repulsion and dipolar interactions, hydrogen bond formation and hydrophobic interactions, as driving forces for membrane association of peptide/proteins has been well established (McLaughlin 1989; McLaughlin and Murray 2005). Peptide-membrane association seems to be mediated by different thermodynamic steps (Figure 5; Jacobs and White 1989; White and Wimley 1999). In the first step, peptide binding is initiated by the electrostatic attraction of the positively charged residues to the anionic membrane. Depending on the peptide charge, and the strength of the membrane surface potential, the electrostatic attraction would increase significantly the protein concentration near the membrane surface, compared with the soluble fraction (Leventis and Silvius 1998; Seelig 2004). The next step implies the transition of the peptide likely into the plane of binding. The exact location of this layer is difficult to define and depends on the hydrophobic/hydrophilic balance of the molecular groups and forces involved. The third step in the binding process is a change of the conformation of the bound peptide (Figure 5). In many cases, peptides are in a random coil conformation in solution but adopt an α-helical conformation when associated to the lipid membrane (e.g., illustrated with melittin and alpha-synuclein, the latest involved in Parkinson disease; Klocek et al. 2009; Lokappa and Ulmer 2011). These conformational transitions also include the random coil-to-β-structure transition (e.g., Alzheimer peptides; Meier and Seelig 2007) and secondary structure reshuffling as recently described for the pore-forming toxin pneumolysin (Tilley et al. 2005). The penetration of the peptide/protein depends on the chemical nature of the lipids, peptides and carbohydrates involved and also on the mechanistic nature of the processes investigated, in which both location and timing of membrane association can be tightly controlled.
Fig. 5.
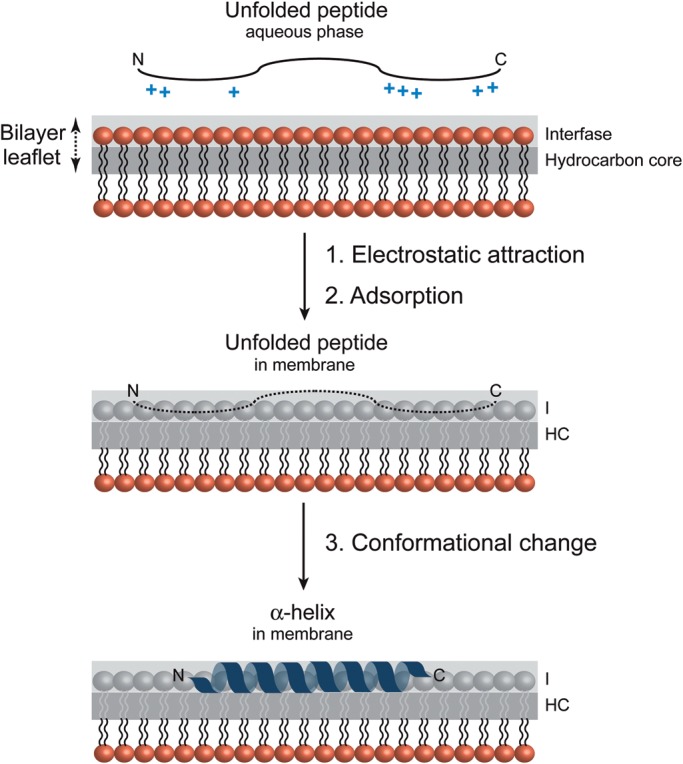
Molecular recognition of a peptide at the membrane surface. Schematic representation of different steps in the process of peptide binding to an anionic phospholipid bilayer. The positively charged peptide is attracted electrostatically to the membrane surface followed by a conformational change to an α-helix (White and Wimley 1999; Seelig 2004).
The crystal structures of TagF (GT2; CDP-Gly, Weidenmaier and Peschel 2008; Lovering et al. 2010), WaaG (GT4; Raetz and Whitfield 2002; Martinez-Fleites et al. 2006), WsaF (GT4; Steiner et al. 2010), WaaC (GT9; Grizot et al. 2006), WaaF (GT-9; pdb code 1PSW), Vpar_0760 (pdb code 3TOV), MurG (GT28; Hu et al. 2003), WaaA (GT30; Schmidt et al. 2012) and GumK (GT70; Barreras et al. 2008), all of which have been proposed to behave similar to the monotopic membrane-associated family of GT-B GTs, clearly display common structural features with AlMGS and AtDGD2. The N-terminal domains are enriched in positively charged residues, mostly in the form of amphiphilic helices, yielding high calculated pI values (Table II; Figure 6). Surface-exposed aromatic hydrophobic residues as tryptophan and phenylalanine are also observed on the N-terminal domain of the crystal structures of TagF, WaaG, WsaF, WbaZ, WaaC, WaaF, MurG and GumK. Interestingly, in TagF, preceding the GT-B region, there are two α-helices that are strong candidates for membrane association and protein oligomerization. The first α-helix (α1; residues 316–331) is highly hydrophobic while the second α-helix (α2; residues 335–344) forms an extremely positively charged environment with neighboring residues (Lovering et al. 2010). In contrast, the Helicobacter pylori α1,3 fucosyltransferase (FucT) is composed of an N-terminal catalytic domain, 2–10 heptad repeats likely involved in protein dimerization and a C-terminal tail which is rich in both basic and hydrophobic residues and would mediate protein–membrane interactions (Sun et al. 2007). These structural features are believed to be involved in anchoring the enzymes to specific lipid environments and to also influence their activities. Moreover, multivariate and bioinformatic sequence analyses identified the occurrence of equivalent structural elements in the N-terminal domain of many structurally uncharacterized membrane-associated GT-B enzymes suggestive of conserved molecular mechanisms of membrane association (Table II; Lind et al. 2007). Major contributions to membrane interaction free energies arise from the desolvation of protein and membrane as they associate and from the electrostatic attraction between protein's basic residues and membranous acid phospholipids (Figure 7; Wimley and White 1996; White and Wimley 1999).
Table II.
Calculated pI of selected membrane-associated GT-B GTs
| GTB-GTa | Length (n° residues) | MW (kDa) | Full pI | NTDb pI | CTDc pI |
|---|---|---|---|---|---|
| TagF | 721 | 85.9 | (313–721) 48.8 kDa 8.14 |
(313–513 + 701–721) 26.5 kDa 9.68 |
(514–700) 22.3 kDa 4.50 |
| WaaA | 353 | 40.7 | (1–353) 40.7 kDa 8.99 |
(1–164 + 336–353) 21.4 kDa 9.65 |
(165–335) 19.3 kDa 8.92 |
| WaaC | 326 | 36.2 | (1–326) 36.2 kDa 8.99 |
(1–162) 18.3 kDa 10.0 |
(163–326) 18.0 kDa 5.67 |
| WaaF | 348 | 39.0 | (1–348) 39.0 kDa 7.15 |
(1–160) 18.4 kDa 9.67 |
(161–348) 20.6 kDa 5.62 |
| WaaG | 374 | 42.3 | (1–374) 42.3 kDa 7.22 |
(1–167 + 358–374) 21.5 kDa 8.35 |
(168–357) 21.1 kDa 5.97 |
| MurG | 355 | 37.8 | (1–355) 37.8 kDa 9.74 |
(1–163 + 341–355) 18.9 kDa 10.38 |
(164–340) 18.9 kDa 7.03 |
| GumK | 400 | 44.4 | (1–400) 44.4 kDa 8.27 |
(1–203 + 361–400) 27.4 kDa 9.22 |
(204–360) 17.0 kDa 6.30 |
| PimA | 386 | 41.2 | (1–386) 41.2 kDa 5.40 |
(1–170 + 348–386) 22.2 kDa 7.11 |
(171–347) 18.9 kDa 4.58 |
aTagF from Staphylococcus epidermidis (pdb code, 3L7I; uniprot code, Q5HLM5_STAEQ), WaaA from Aquifex aeolicus (2XCI, KDTA_AQUAE), WaaC from E. coli (2GT1, E2QGX8_ECOLX), WaaF from E. coli (1PSW, N2I599_ECOLX), WaaG from E. coli (2IW1, RFAG_ECOLI), MurG from E. coli (1F0K, MURG_ECOBW), GumK from Xanthomonas campestris (2Q6V, GUMK_XANCP), PimA from M. smegmatis (2GEK, PIMA_MYCS2).
bNTD, N-terminal domain.
cCTD, C-terminal domain.
Fig. 6.
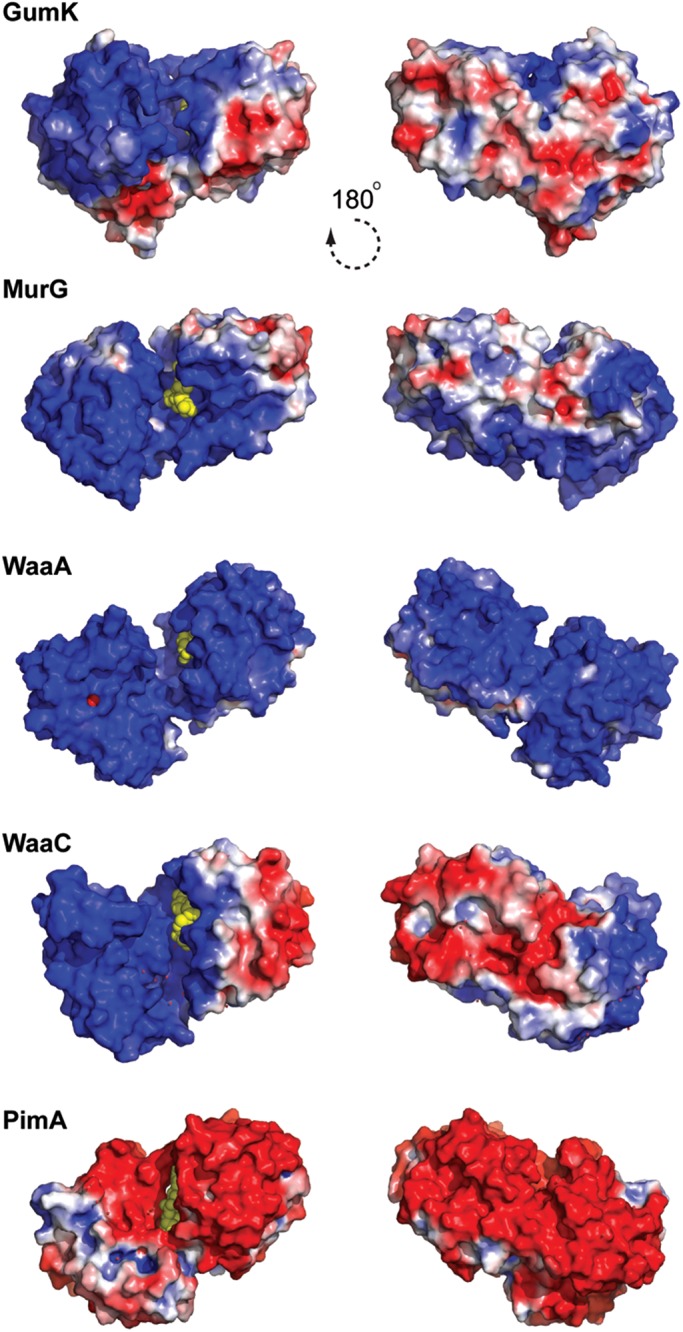
Electrostatic surface potential representation of membrane-associated GT-B GTs, as visualized in GumK, MurG, WaaA, WaaC and PimA.
Fig. 7.
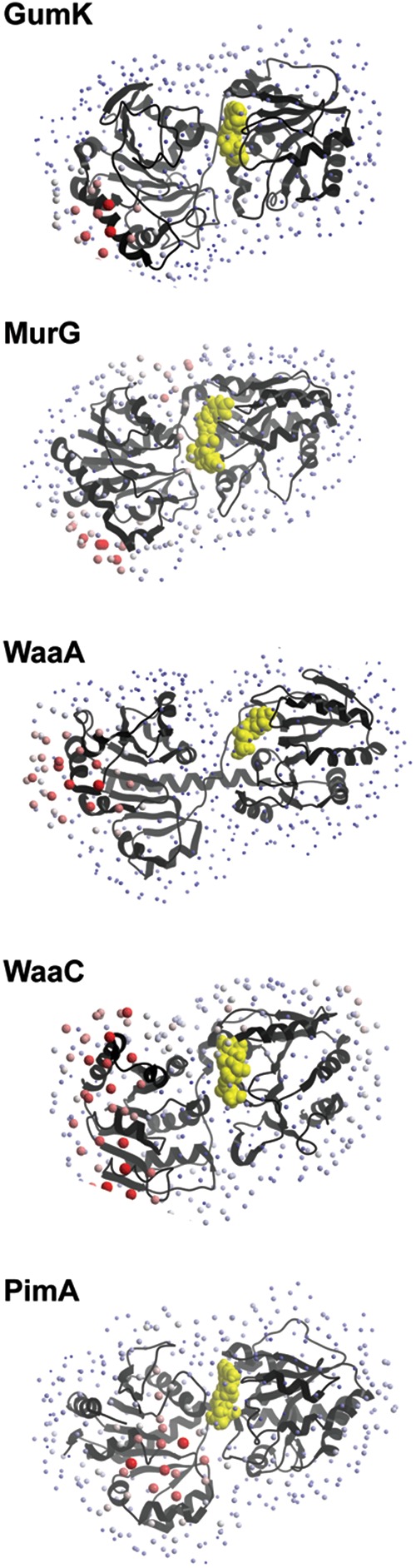
Calculation of the desolvation energy values derived from octanol/water experiments. The hotspots indicate areas with the most favorable energy change upon binding, likely to be buried in the membrane. The largest hotspots correspond to energy values lower than −15.0 kcal mol−1, medium-sized spots represent values between −15 and −10 kcal mol−1 and the smallest points represent values higher than −10 kcal mol−1.
Peripheral GT-B GTs
The membrane association and dissociation rate constants of many GT-B GTs remain largely unknown, making their unambiguous classification as monotopic or peripheral membrane-associated enzymes difficult. Consequently, the determination of the protein sub-cellular localization became, in most of the cases, the sole criteria to discriminate between both groups of enzymes. The phosphatidylinositol mannosyltransferase A, which has been shown to co-localize both in the membrane and in cytosolic fractions, represents a clear example of a peripheral membrane-associated GT-B GT (PimA; Korduláková et al. 2002; Guerin et al. 2009). PimA is an essential enzyme for mycobacterial growth that initiates the biosynthetic pathway of key structural elements and virulence factors of Mycobacterium tuberculosis, the phosphatidyl-myo-inositol mannosides (PIM), lipomannan and lipoarabinomannan (Kaur et al. 2009; Guerin et al. 2010; Morita et al. 2011). The enzyme catalyzes the transfer of a mannose residue from GDP-Man to the 2-position of inositol phosphate ring in phosphatidyl-myo-inositol (PI) to form phosphatidyl-myo-inositol monomannoside (PIM1) on the cytoplasmic side of the plasma membrane (Guerin et al. 2009). The crystal structure of PimA in complex with its donor sugar substrate pointed to a region located in the N-terminal domain and adjacent to the two-domain cleft, as potentially mediating protein–membrane interactions. Specifically, the interfacial-binding surface contains a solvent-exposed hydrophobic patch close to a cluster of basic residues in an amphiphatic α-helix and to the connecting loop β3-α2, which is disordered in the lipid-free structure (Guerin et al. 2007). Supporting this notion, a PimA mutant in which residues Arg77, Lys78, Lys80 and Lys81 on α-helix 2 were substituted by serine completely inactivated the enzyme and drastically impaired the ability of the protein to bind PI (Guerin et al. 2007; Guerin et al. 2009). It is worth noting that the positively charged cluster is far from the catalytic center and exposed to solvent, and therefore, it is not expected to interfere with the catalytic machinery. Moreover, the involvement of this cluster in protein–membrane interactions is consistent with the observed activity enhancement of PimA in the presence of nonsubstrate anionic phospholipids including 1,2-dipalmitoyl-sn-glycero-3-phosphate or cardiolipin (Guerin et al. 2007). It may also explain previous results showing that a salt wash of mycobacterial plasma membranes significantly reduced the biosynthesis of PIM1 (Morita et al. 2005).
However, peripheral GT-B GTs seem to have acquired different strategies to associate to the membrane, according to the specific functions they perform inside the cell (Figure 8; Lemmon 2008; Moravcevic et al. 2012). In this regard, it is interesting to mention the case of Alg13/Alg14, an heterodimeric enzyme involved in the biosynthesis of N-glycans in Saccharomyces cerevisiae (Gao et al. 2005). Alg13/Alg14 mediates the second step of the pathway by transferring a GlcNAc residue from UDP-GlcNAc donor to the Dol-PP-GlcNAc acceptor. Alg14 is a bitopic membrane-associated protein, with a cytosolic-predicted Rossmann-like fold domain. According to the NMR solution structure, Alg13 adopts a nonconventional Rossmann-fold. Titration of Alg13 with UDP-GlcNAc demonstrated that Alg13 interacts with the sugar donor through residues located on the C-terminal half of the protein. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form the bipartite enzyme, where the C-terminal region seems to modulate Alg13/Alg14 protein–protein interactions (Gao et al. 2007). Another good example is the plant sucrose synthase (SUS). SUS participates in the modulation of sucrose flux by rapidly altering its cellular location from the cytosol to other sites including with various organelle membranes (Subbaiah et al. 2006). However, the molecular mechanisms by which SUS binds to its cellular targets, the structural aspects controlling SUS partitioning and the structural impact of partitioning on its catalytic function are currently unknown. In Zea mays, the phosphorylation of the SUS1 isoform at the Ser-15 residue increases membrane association and catalytic activity at acidic pH in Zea mays (Hardin et al. 2004). Interestingly, phosphorylation of SUS1 modifies the oligomerization state of SUS1, from dimers to tetramers. The recently solved crystal structure of SUS1 from A. thaliana suggests a model that explains how SUS catalysis and the interaction with its cellular targets might be regulated (Zheng et al. 2011). A conserved sequence surrounding the equivalent Ser-13 is positively charged (RXHSX(R/K)ER), whereas the neighboring helix α8 has both anionic and cationic patches (LKRAEEYL) most probably altering the electrostatic environment in this region.
Fig. 8.
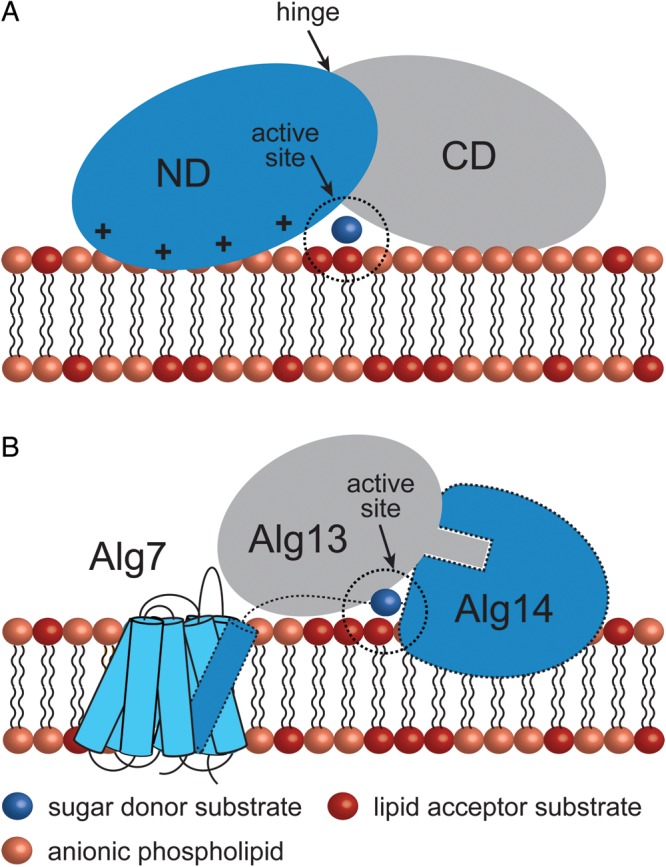
Peripheral GT-B GTs adopt different strategies for membrane association. (A) The mycobacterial mannosyltransferase PimA interacts with the phospholipid bilayer by a combination of positively charged and hydrophobic residues exposed on the N-terminal domain of the protein. An important conformational change is expected to occur during/after protein–membrane interaction (Guerin et al. 2007; Guerin et al. 2009; Giganti et al. 2013). (B) A model for the Alg7/13/14 complex formation. This functional multienzyme complex catalyzes the first two steps of lipid-linked oligosaccharide on the cytosolic face of the ER membrane. Alg7, a politopic protein, which transfers a GlcNAc-phosphate to dolichol phosphate, interacts with the transmembrane α-helix of the bitopic GT-GT Alg14 (Lu et al. 2012). Biochemical and structural data support a model in which Alg13-Ag14 interaction is mediated by residues located on the C-terminal α-helix of Alg13 and C-terminal amino acids of Alg14 to form a dimeric Alg13/14 (Wang et al. 2008).
Nucleotide sugar donor-binding site
The nucleotide mono- or diphospho-sugar-binding sites essentially comprise a hydrophobic pocket and a conserved α/β/α motif located on the more conserved C-terminal domain of the enzymes, and a region constituted by the β1-α1 loop and the top of the α1 on the N-terminal domain, with all regions facing the interdomain cleft (Hu and Walker 2002; Figure 9A and B, Table I). The nucleobase heterocycle accommodates into the hydrophobic pocket where it makes electrostatic interactions and hydrogen bonds with the GT. These noncovalent interactions account for the nucleobase selectivity of this family of enzymes (Figure 9C). The first α-helix (αF) of the α/β/α motif (αF/β/αS) mainly interacts with the O2 and O3 hydroxyl groups of the ribose ring, while the second α-helix (αS) and the β-α2 connecting loop makes key contacts with the sugar ring. It is worth noting that α-helices possess a permanent dipole moment with a positively charged N-terminus and a negatively charged C-terminus. As a consequence, it has been observed that α-helices favor the interaction with ionic species in which the first and second turn seem to be critical for the electrostatic interaction to occur (Aqvist et al. 1991). In that sense, it is believed that in GT-B GTs the negatively charged pyrophosphate group is stabilized by α1 (N-terminal domain) and αS (C-terminal domain) through helix dipole effect (Figure 9C; Hol et al. 1978; Hu and Walker 2002). In many cases, the α- and β-phosphates are further stabilized by positively charged side chain residues (Arg and/or Lys; Figure 9C; Lairson et al. 2008) and/or the β1-α1 loop located in the N-terminal domain. This loop is known to undergo significant conformational changes upon sugar-nucleotide binding in many GT-B GTs (Buschiazzo et al. 2004; Vetting et al. 2008; Sheng et al. 2009).
Fig. 9.
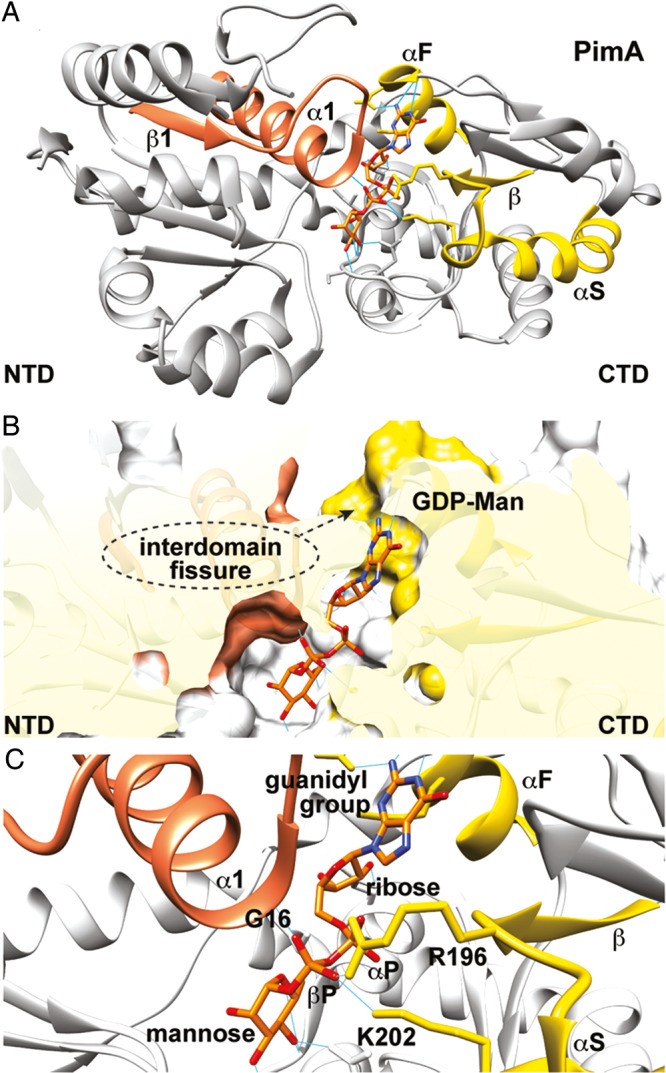
The nucleotide sugar donor-binding site. (A) Overall structure of the phosphatidylinositol mannosyltransferase PimA (PM, GT4) from M. smegmatis showing the β1-α1 motif (residues Met1 to Ala31) in orange and the α/β/α motif (αF/β/αS; residues Asp252 to Gly287) in yellow (Guerin et al. 2007). (B) Surface representation of the active site of PimA showing the hydrophobic binding pocket and Asp253 that account for nucleoside heterocycle specificity (Guerin et al. 2007). (C) The negatively charged pyrophosphate group is stabilized by α1 (Gly15 to Ala31 at the N-terminal domain) and αS (Gly277 to Gly287 at the C-terminal domain) through helix dipole effect (Aqvist et al. 1991; Hol et al. 1978; Hu and Walker 2002). The α- and β-phosphates are further stabilized by two positively charged residues, Arg196 and Lys202, and the α1-β1 loop.
Sugar acceptor-binding site
To date, no direct structural information is available for any integral membrane-associated GT-B GT, including monotopic, bitopic and polytopic proteins, in the presence of their acceptor substrates. However, the acceptor-binding site is expected to be located in the N-terminal domain as occur with the soluble enzymes (Lairson et al. 2008). Supporting this notion, the crystal structure of WaaG revealed a deep and open cavity in which the sugar moiety of the donor is found to cover the deepest point of its floor. This pocket encloses an area of ∼940 Å, able to accommodate the first two sugar moieties of the acceptor LPS and eventually the glycosyl ramification on the first of these residues. The walls of the cavity are lined by four tyrosine residues and with various lysine and arginine residues pointing directly to the cavity, in line with the generic signature of carbohydrate-binding sites (Martinez-Fleites et al. 2006). Similarly, the N-terminal domain of WaaA exhibits a depression that extends from the putative membrane-association site to the central groove between the two domains. A PEG molecule has been detected in the |Fo| − |Fc| difference density map, strongly suggesting that this particular region may serve as the acceptor-substrate-binding site for the lipid A precursor (Schmidt et al. 2012). A close inspection of the monotopic AviGT4, WaaA, WaaG, WsaF, WbaZ, WaaC, WaaF and MurG, and the bitopic Fut8 crystal structures also revealed the presence of a cavity, of variable shape and volume and located nearby the sugar donor-binding site, suitable for binding the acceptor molecules. However, to improve the efficient glycosylation of membrane-bound acceptors such as lipids/glycolipids, a close proximity between the lipid bilayer and the acceptor-binding sites is likely to be required. Structural comparison of the N-terminal domains of the monotopic GT-B structures currently available showed that the amphipathic α-helices display very different orientations, with all of them being exposed to the solvent. Thus, important conformational changes are expected to occur in the proteins after membrane/acceptor substrates association.
The crystal structure of SUS1, a peripheral membrane-associated GTB GT, has been recently solved in the presence of UDP and its sugar acceptor fructose at 2.85 Å resolution (Zheng et al. 2011). The enzyme transfers a glucose residue to the 2-position of fructose to form sucrose in plants. The fructose is firmly bound in the β-furanose form within a pocket formed exclusively by residues from the N-terminal domain of the protein. Along the same line, the molecular architecture of the lipid acceptor-binding site of PimA has been proposed to occur on the N-terminal domain by using in silico molecular docking approaches (Guerin et al. 2009). The inositol phosphate group accommodates into a well-defined pocket with its O2 atom favorably positioned to receive the mannosyl residue from GDP-Man. A similar scenario has been proposed for PimB’, the second enzyme in the PIM biosynthetic pathway (Batt et al. 2010). However, the interplay between the membrane and acceptor-binding site in peripheral membrane-associated GT-B GTs seems to be diverse at the molecular level.
Conformational changes
To achieve the enzyme-transition state complex, a spatial rearrangement of the active site is very often required, highlighting the importance of protein dynamics and conformational changes in substrate recognition (Hammes-Schiffer and Benkovic 2006; Schramm 2011). Specifically, protein conformational changes not only involve local reorganization of flexible loops and side-chain residues, but also, in many cases, domain motions and protein oligomerization events. Moreover, as a consequence of protein–protein interactions, post-translational modifications and noncovalent associations with small-molecule inhibitors or activators, conformational dynamics critically modulates enzyme catalysis.
The “open”-to-“closed” motion
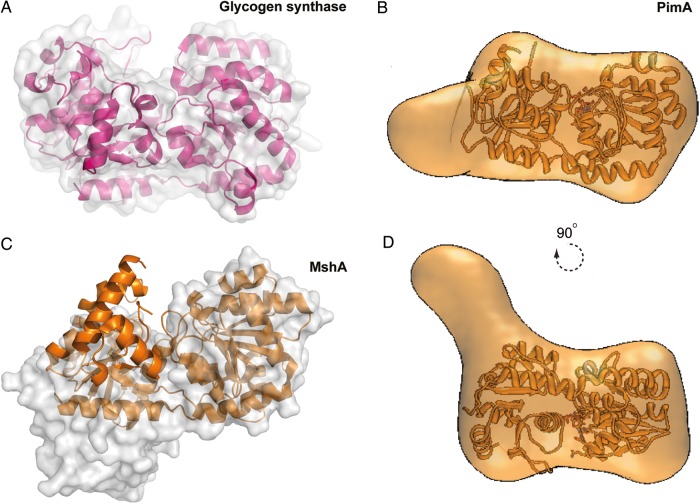
Structural, biochemical and biophysical studies have demonstrated that members of the GT-B superfamily, including soluble and membrane-associated enzymes, can adopt both open and closed forms (Buschiazzo et al. 2004; Guerin et al. 2009; Giganti et al. 2013). It has been proposed that nucleotide sugar donor binding triggers a closure movement that involves the rotation of the N- and C-terminal domains, which brings together critical residues making up a functionally competent catalytic center. This conformational flexibility can be dramatic, as is illustrated by the structures of the ligand-free and UDP-GlcNAc bound complex of MshA from Corynebacterium glutamicum in which the relative orientation of the N- and C-terminal domains involves a rotational movement of 97° (Vetting et al. 2008). Other very good examples are given by the glycogen synthase (Buschiazzo et al. 2004; Sheng et al. 2009) and the monotopic MurG (Hu et al. 2003; Figure 10A).
Fig. 10.
Conformational changes on GT-B GTs. (A) Structural comparison between the “open” (molecular surface representation) and “closed” (schematic cartoon) states of glycogen synthase (Sheng et al. 2009) and (C) MshA from C. glutamicum (Vetting et al. 2008). (B, D) Average low-resolution structure of PimA-GDP complex with the high-resolution crystal structure of PimA-GDP complex fitted by rigid body docking (Giganti et al. 2013).
However, perhaps the most well-documented studies about the conformational changes a GT-B enzyme undergoes upon substrate binding corresponds to the peripheral membrane-associated PimA from mycobacteria. The binding of the sugar nucleotide donor GDP-Man induces an interdomain rearrangement from an “open” to a “closed” state that stabilizes the enzyme. The interaction of PimA with the β-phosphate of GDP-Man was essential for this conformational change to occur (Guerin et al. 2009). Several lines of experimental evidence strongly support this notion. First, the crystal structures of the PimA-GDP and PimA-GDP-Man complexes revealed that GDP-Man is buried at the N- and C-terminal domains interface, with residues Gly16, Arg196 and Lys202 stacking the β-phosphate (Guerin et al. 2007). It is worth mentioning that the α-PO4 of the donor substrate does not interact with any particular residue from the enzyme. Secondly, PimA-limited proteolysis studies confirmed that the enzyme was rapidly degraded after incubation with elastase. N-terminal sequencing of the two predominant species of 23 and 15 kDa revealed two exposed sites located in α9, involved in GDP-Man recognition, and the connecting loop β7-β8 at the junction between N- and C-terminal domains. GDP or GDP-Man, but not guanosine where the α-PO4 and β-PO4 are missing, protected PimA from the action of the protease even after 90 min of incubation. The “open-to-closed” motion was further monitored in the absence and presence of GDP and guanosine by sedimentation velocity analytical ultracentrifugation studies of pure PimA (Guerin et al. 2009). PimA sedimented as a single homogeneous species with an average sedimentation coefficient of 3.22 s, which is consistent with an asymmetric monomeric protein. Upon addition of equimolar GDP, the sedimentation coefficient increased to 3.53 s, indicating the formation of a more symmetrical and compact structure. As expected, the addition of guanosine did not significantly affect the sedimentation coefficient value of the PimA. Moreover, ITC measurements revealed that guanosine binds to PimA with a binding constant ∼103-fold smaller than that of GDP. The GDP binding also produces a stabilizing effect on PimA characterized by an increment of ∼3.5°C in the melting temperature value as observed by differential scanning calorimetry and circular dicroism (Guerin et al. 2009). More recently, the ab initio low-resolution envelopes obtained from small-angle X-ray scattering of the unliganded PimA and the PimA-GDP complexed forms clearly demonstrate that the “open” and “closed” conformations of the GT-B enzyme are largely present in solution (Giganti et al. 2013; Figure 10B). Specifically, the radii of gyration (Rg) obtained for PimA apo and the PimA-GDP complex revealed a clear reduction in Rg (ΔRg) of −1.0 (1) Å. In summary, the sugar nucleotide donor induces the closure movement of PimA, with a clear increment of the sedimentation coefficient value which is correlated with a reduction of the radii of gyration, and accompanied by a markedly stabilization of the enzyme.
In contrast, the binding of the acceptor substrate PI to the enzyme had an effect opposite to that observed for the sugar nucleotide donor. When incubated with PI, PimA became highly sensitive to elastase, even in the presence of GDP, indicating that PI triggers a yet significant conformational change that modifies the closed GDP-Man-induced conformation (Guerin et al. 2009). Furthermore, analytical ultracentrifugation experiments demonstrated that the addition of PI to the enzyme resulted in a significant change in the sedimentation coefficient values of both unliganded PimA and PimA-GDP complex consistent with the formation of less compact structures, which correlates with a reduction of the melting temperature by 1.5 and 0.4°C, respectively, in agreement with the limited proteolysis experiments. Interestingly, single-molecule force spectroscopy indicated that PimA unfolds following heterogeneous multiple-step mechanical unfolding pathways at low force, akin to molten globule states. Small-angle X-ray scattering data revealed that PimA experiences remarkable flexibility that undoubtedly corresponds to the N-terminal domain, which has been proposed to participate in lipid acceptor and protein–membrane interactions (Guerin et al. 2009; Giganti et al. 2013). Altogether, the experimental data support a model wherein the flexibility and conformational transitions confer the adaptability of PimA to the donor and acceptor substrates, and to the membrane, which seems to be of importance during catalysis.
Future challenges
Substantial advances have been made over the last 10 years in understanding the membrane-associated GT-B of GTs, mostly due to the 3D structural and biophysical characterization of several monotopic and peripheral family members. The emerging importance of protein dynamics in membrane-associated GT-B anticipates exciting times in the field because considerable challenges remain to be overcome to fully understand the mode of action of these enzymes. Among these challenges are: (i) understanding the dynamics and conformational changes in GT-B GTs during membrane association and substrate binding; (ii) elucidating how substrates and products diffuses between the hydrophobic environment of the membrane and the catalytic center of the enzyme; (iii) determining the molecular structures of membrane-associated GT-B GTs in complex with lipid acceptors; (iv) understanding how the substrate-binding affinities and the catalytic efficiencies of these enzymes can be modulated by post-translational modifications, membrane composition or protein mobility (Ramadurai et al. 2009); (v) investigating the physical properties of the membranes (e.g., thickness, curvature and phospholipid packing) and their possible influence on the catalytic properties of GTs or yet (vi) discovering new potent GT inhibitors as research tools to gain further insights into the molecular mechanisms of membrane association and catalysis, with potential applications in drug discovery.
Funding
This work was supported by European Commission Contract HEALTH-F3–2011-260872 (More Medicines for Tuberculosis), the Spanish Ministry of Science and Innovation Contract SAF2010-19096, the National Institutes of Health/National Institute of Allergy and Infectious Diseases grant AI064798, IKERBASQUE, the Basque Foundation for Science and the Basque Government.
Conflict of interest
None declared.
Abbreviations
BGT, β-glucosyltransferase; CAZy, Carbohydrate-Active enZymes; CL, cardiolipin; DAG, sn-1,2-diacylglycerol; DOPG, 1,2-dioleoyl-phosphatidylglycerol; FucT, α1,3 fucosyltransferase; GlcNAc, N-acetylglucosaminyltransferase; GTA, N-acetyl-galactosaminyltransferase; GTs, glycosyltransferases; MshA, UDP-GlcNAc:inositol-P N-acetylglucosaminyltransferase; MurG, UDP-GlcNAc: N-acetylmuramyl-(pentapeptide)-PP-C55 N-acetyl-glucosaminyltransferase; NMR, nuclear magnetic resonance; PG, phosphatidylglycerol; PI, phosphatidyl-myo-inositol; PIM, phosphatidyl-myo-inositol mannosides; PIM1, phosphatidyl-myo-inositol monomannoside; PimA, GDP-Man:phosphatidylinositol mannosyltransferase; SPR, surface plasmon resonance; SUS, sucrose synthase; UDP, uridine diphosphate; MurNAc, N-acetylmuramic acid; GDP, guanosine diphosphate; ITC, isothermal titration calorimetry.
Acknowledgments
We gratefully acknowledge the valuable scientific discussions with Prof. Åke Wieslander (Department of Biochemistry and Biophysics, Stockholm University) and Prof. Antoni Planas (IQS, Barcelona, Spain) and all members of the Structural Glycobiology Group (Spain). We thank Michael Nilges (Institut Pasteur, Paris, France), who provided the ICM software for the calculation of the desolvation energy values. D.A.J. and D.G. acknowledge the support from the Fundación Biofísica Bizkaia (Spain).
References
- Abdian PL, Lellouch AC, Gautier C, Ielpi L, Geremia RA. Identification of essential amino acids in the bacterial alpha-mannosyltransferase AceA. J Biol Chem. 2000;275:40568–40575. doi: 10.1074/jbc.M007496200. doi:10.1074/jbc.M007496200. [DOI] [PubMed] [Google Scholar]
- Andersen OS, Koeppe RE., II Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. doi:10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- Andersson AS, Rilfors L, Bergqvist M, Persson S, Lindblom G. New aspects on membrane lipid regulation in Acholeplasma laidlawii A and phase equilibria of monoacyl-diglucosyldiacylglycerol. Biochemistry. 1996;35:11119–11130. doi: 10.1021/bi960561w. doi:10.1021/bi960561w. [DOI] [PubMed] [Google Scholar]
- Andrés E, Biarnés X, Faijes M, Planas A. Bacterial glycoglycerolipid synthases: Processive and non-processive glycosyltransferases in mycoplasma. Biocatal Biotransfor. 2012;30:274–287. doi:10.3109/10242422.2012.674733. [Google Scholar]
- Ardévol A, Rovira C. The molecular mechanism of enzymatic glycosyl transfer with retention of configuration: Evidence for a short-lived oxocarbenium-like species. Angew Chem Int Ed Engl. 2011;50:10897–10901. doi: 10.1002/anie.201104623. doi:10.1002/anie.201104623. [DOI] [PubMed] [Google Scholar]
- Ariöz C, Ye W, Bakali A, Ge C, Götzke H, Liebau J, Barth A, Wieslander Å, Mäler L. Anionic lipid binding to the foreign protein MGS provides a tight coupling of phospholipid synthesis and protein overexpression in Escherichia coli. Biochemistry. 2013;52:5533–5544. doi: 10.1021/bi400616n. doi:10.1021/bi400616n. [DOI] [PubMed] [Google Scholar]
- Artymiuk PJ, Rice DW, Poirrette AR, Willett P. β-glucosyltransferase and phosphorylase reveal their common theme. Nat Struct Biol. 1995;2:117–120. doi: 10.1038/nsb0295-117. doi:10.1038/nsb0295-117. [DOI] [PubMed] [Google Scholar]
- Aqvist J, Luecke H, Quiocho FA, Warshel A. Dipoles localized at helix termini of proteins stabilize charges. Proc Natl Acad Sci USA. 1991;88:2026–2030. doi: 10.1073/pnas.88.5.2026. doi:10.1073/pnas.88.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balali-Mood K, Bond PJ, Sansom MS. Interaction of monotopic membrane enzymes with a lipid bilayer: A coarse-grained MD simulation study. Biochemistry. 2009;48:2135–2145. doi: 10.1021/bi8017398. doi:10.1021/bi8017398. [DOI] [PubMed] [Google Scholar]
- Barford D, Johnson LN. The allosteric transition of glycogen phosphorylase. Nature. 1989;340:609–616. doi: 10.1038/340609a0. doi:10.1038/340609a0. [DOI] [PubMed] [Google Scholar]
- Barreras M, Salinas SR, Abdian PL, Kampel MA, Ielpi L. Structure and mechanism of GumK, a membrane associated glucuronosyltransferase. J Biol Chem. 2008;283:25027–25035. doi: 10.1074/jbc.M801227200. doi:10.1074/jbc.M801227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran S, Roach PJ, Depaoli-Roach AA, Hurley TD. Structural basis for glucose-6-phosphate activation of glycogen synthase. Proc Natl Acad Sci USA. 2010;107:17563–17568. doi: 10.1073/pnas.1006340107. doi:10.1073/pnas.1006340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt SM, Jabeen T, Mishra AK, Veerapen N, Krumbach K, Eggeling L, Besra GS, Fütterer K. Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: Structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB. J Biol Chem. 2010;285:37741–37752. doi: 10.1074/jbc.M110.165407. doi:10.1074/jbc.M110.165407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Ohta H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem. 2005;280:2397–2400. doi: 10.1074/jbc.R400032200. doi:10.1074/jbc.R400032200. [DOI] [PubMed] [Google Scholar]
- Berg S, Edman M, Li L, Wikstrom M, Wieslander Å. Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J Biol Chem. 2001;276:22056–22063. doi: 10.1074/jbc.M102576200. doi:10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- Berg S, Kaur D, Jackson M, Brennan PJ. The glycosyltransferases of Mycobacterium tuberculosis—Roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology. 2007;17:35–56R. doi: 10.1093/glycob/cwm010. Review doi:10.1093/glycob/cwm010. [DOI] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. doi:10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. Review doi:10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. doi:10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- Brown K, Vial SC, Dedi N, Westcott J, Scally S, Bugg TD, Charlton PA, Cheetham GM. Crystal structure of the Pseudomonas aeruginosa MurG:UDP-GlcNAc substrate complex. Protein Pept Lett. 2013;20:1002–1008. doi: 10.2174/0929866511320090006. doi:10.2174/0929866511320090006. [DOI] [PubMed] [Google Scholar]
- Buschiazzo A, Ugalde JE, Guerin ME, Shepard W, Ugalde RA, Alzari PM. Crystal structure of glycogen synthase: Homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. doi:10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Davies GJ. Structure of the nucleotidediphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38:6380–6385. doi: 10.1021/bi990270y. doi:10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- Chen L, Men H, Ha S, Ye X, Brunner L, Hu Y, Walker S. Intrinsic lipid preferentes and kinetic mechanism of Escherichia coli MurG. Biochemistry. 2002;41:6824–6833. doi: 10.1021/bi0256678. doi:10.1021/bi0256678. [DOI] [PubMed] [Google Scholar]
- Chung BC, Zhao J, Gillespie RA, Kwon D, Guan Z, Hong J, Zhou P, Lee SY. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. doi:10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. doi:10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Nordstrom S, Karlsson OP, Mannock DA, McElhaney RN, Wieslander Å. Efficient modulation of glucolipid enzyme activities in membranes of Acholeplasma laidlawii by the type of lipids in the bilayer matrix. Biochemistry. 1995;34:13381–13389. doi: 10.1021/bi00041a015. doi:10.1021/bi00041a015. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Planas A, Rovira C. Conformational analyses of the reaction coordinate of glycosidases. Acc Chem Res. 2012;45:308–316. doi: 10.1021/ar2001765. doi:10.1021/ar2001765. [DOI] [PubMed] [Google Scholar]
- Dormann P, Benning C. Galactolipids rule in seed plants. Trends Plant Sci. 2002;7:112–118. doi: 10.1016/s1360-1385(01)02216-6. doi:10.1016/S1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- Edman M, Berg S, Storm P, Wikström M, Vikström S, Ohman A, Wieslander Å. Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J Biol Chem. 2003;278:8420–8428. doi: 10.1074/jbc.M211492200. doi:10.1074/jbc.M211492200. [DOI] [PubMed] [Google Scholar]
- Elofsson A, von Heijne G. Membrane protein structure: Prediction versus reality. Ann Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. doi:10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. doi:10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- Eriksson HM, Wessman P, Ge C, Edwards K, Wieslander Å. Massive formation of intracellular membrane vesicles in Escherichia coli by a monotopic membrane-bound lipid glycosyltransferase. J Biol Chem. 2009;284:33904–33914. doi: 10.1074/jbc.M109.021618. doi:10.1074/jbc.M109.021618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Mattevi A. Enzymes without borders: Mobilizing substrates, delivering products. Science. 2008;321:213–216. doi: 10.1126/science.1151118. doi:10.1126/science.1151118. [DOI] [PubMed] [Google Scholar]
- Frantom PA, Coward JK, Blanchard JS. UDP-(5F)-GlcNAc acts as a show-binding inhibitor of MshA, a retaining glycosyltransferase. J Am Chem Soc. 2010;132:6626–6627. doi: 10.1021/ja101231a. doi:10.1021/ja101231a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Moriyama S, Miura N, Nishimura SI. C-termini of both alg13 and alg14 proteins are required for formation of the alg13/alg14 complex. Glycobiology. 2007;17:1277. [Google Scholar]
- Gao XD, Tachikawa H, Sato T, Jigami Y, Dean N. Alg14 recruits alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J Biol Chem. 2005;280:36254–36262. doi: 10.1074/jbc.M507569200. doi:10.1074/jbc.M507569200. [DOI] [PubMed] [Google Scholar]
- Ge C. Sweden: Department of Biochemistry and Biophysics, Stockholm University; 2011. Property-controlling enzymes at the membrane interface. Ph.D. dissertation. [Google Scholar]
- Ge C, Georgiev A, Öhman A, Wieslander Å, Kelly AA. Tryptophan residues promote membrane association for a plant lipid glycosyltransferase involved in phosphate stress. J Biol Chem. 2011;286:6669–6684. doi: 10.1074/jbc.M110.138495. doi:10.1074/jbc.M110.138495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson RP, Turkenburg JP, Charnock SJ, Lloyd R, Davies GJ. Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase OtsA. Chem Biol. 2002;9:1337–1346. doi: 10.1016/s1074-5521(02)00292-2. doi:10.1016/S1074-5521(02)00292-2. [DOI] [PubMed] [Google Scholar]
- Giganti D, Alegre-Cebollada J, Urresti S, Albesa-Jove D, Rodrigo-Unzueta A, Comino N, Kachala M, Lopez-Fernandez S, Svergun DI, Fernandez JM, et al. Conformational plasticity of the essential membrane associated mannosyltransferase PimA from mycobacteria. J Biol Chem. 2013;288:29797–29808. doi: 10.1074/jbc.M113.462705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Cunningham AM, Watson DC, Martin A, Richards JC, Wakarchuk WW. Characterization of a recombinant Neisseria meningitidis alpha-2,3-sialyltransferase and its acceptor specificity. Eur J Biochem. 1997;249:187–194. doi: 10.1111/j.1432-1033.1997.t01-1-00187.x. doi:10.1111/j.1432-1033.1997.t01-1-00187.x. [DOI] [PubMed] [Google Scholar]
- Gomez H, Polyak I, Thiel W, Lluch JM, Masgrau L. Retaining glycosyltransferase mechanism studied by QM/MM methods: Lipopoly-saccharyl-α-1,4-galactosyltransferase C transfers α-galactose via an oxocarbenium ion-like transition state. J Am Chem Soc. 2012;134:4743–4752. doi: 10.1021/ja210490f. doi:10.1021/ja210490f. [DOI] [PubMed] [Google Scholar]
- Grizot S, Salem M, Vongsouthi V, Durand L, Moreau F, Dohi H, Vincent S, Escaich S, Ducruix A. Structure of the Escherichia coli heptosyltransferase WaaC: Binary complexes with ADP and ADP-2-deoxy-2-fluoro heptose. J Mol Biol. 2006;363:383–394. doi: 10.1016/j.jmb.2006.07.057. doi:10.1016/j.jmb.2006.07.057. [DOI] [PubMed] [Google Scholar]
- Guerin ME, Kaur D, Somashekar BS, Gibbs S, Gest P, Chatterjee D, Brennan PJ, Jackson M. New insights into the early steps of phosphatidylinositol mannoside biosynthesis in mycobacteria: PimB’ is an essential enzyme of Mycobacterium smegmatis. J Biol Chem. 2009;284:25687–25696. doi: 10.1074/jbc.M109.030593. doi:10.1074/jbc.M109.030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin ME, Korduláková J, Alzari PM, Brennan PJ, Jackson M. Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J Biol Chem. 2010;285:33577–33583. doi: 10.1074/jbc.R110.168328. Review doi:10.1074/jbc.R110.168328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin ME, Kordulakova J, Schaeffer F, Svetlikova Z, Buschiazzo A, Giganti D, Gicquel B, Mikusova K, Jackson M, Alzari PM. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J Biol Chem. 2007;282:20705–20714. doi: 10.1074/jbc.M702087200. doi:10.1074/jbc.M702087200. [DOI] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. doi:10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. doi:10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- Hardin SC, Winter H, Huber SC. Phosphorylation of the amino terminus of maize sucrose synthase in relation to membrane association and enzyme activity. Plant Phys. 2004;134:1427–1438. doi: 10.1104/pp.103.036780. doi:10.1104/pp.103.036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. Review doi:10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. Review doi:10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hol WG, van Duijnen PT, Berendsen HJ. The alpha-helix dipole and the properties of proteins. Nature. 1978;273:443–446. doi: 10.1038/273443a0. doi:10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci USA. 2003;100:845–849. doi: 10.1073/pnas.0235749100. doi:10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. Review doi:10.1016/S1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- Ihara H, Ikeda Y, Toma S, Wang X, Suzuki T, Gu J, Miyoshi E, Tsukihara T, Honke K, Matsumoto A, et al. Crystal structure of mammalian alpha1,6-fucosyltransferase, FUT8. Glycobiology. 2007;17:455–466. doi: 10.1093/glycob/cwl079. doi:10.1093/glycob/cwl079. [DOI] [PubMed] [Google Scholar]
- Iwatani T, Okino N, Sakakura M, Kajiwara H, Takakura Y, Kimura M, Ito M, Yamamoto T, Kakuta Y. Crystal structure of alpha/beta-galactoside alpha2,3-sialyltransferase from a luminous marine bacterium, Photobacter-ium phosphoreum. Febs Lett. 2009;583:2083–2087. doi: 10.1016/j.febslet.2009.05.032. doi:10.1016/j.febslet.2009.05.032. [DOI] [PubMed] [Google Scholar]
- Jacobs RE, White SH. The nature of the hydrophobic binding of small peptides at the bilayer interface: Implications for the insertion of transbilayer helices. Biochemistry. 1989;28:3421–3437. doi: 10.1021/bi00434a042. doi:10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- Kakuda S, Shiba T, Ishiguro M, Tagawa H, Oka S, Kajihara Y, Kawasaki T, Wakatsuki S, Kato R. Structural basis for acceptor substrate recognition of a human glucuronyltransferase, GlcAT-P, an enzyme critical in the biosynthesis of the carbohydrate epitope HNK-1. J Biol Chem. 2004;279:22693–22703. doi: 10.1074/jbc.M400622200. doi:10.1074/jbc.M400622200. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Okino N, Kajiwara H, Ichikawa M, Takakura Y, Ito M, Yamamoto T. Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 alpha2,6- sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: Catalytic mechanism and substrate recognition. Glycobiology. 2008;18:66–73. doi: 10.1093/glycob/cwm119. doi:10.1093/glycob/cwm119. [DOI] [PubMed] [Google Scholar]
- Karlsson OP, Dahlqvist A, Vikstrom S, Wieslander Å. Lipid dependence and basic kinetics of the purified 1,2-diacylglycerol 3-glucosyltransferase from membranes of Acholeplasma laidlawii. J Biol Chem. 1997;272:929–936. doi: 10.1074/jbc.272.2.929. doi:10.1074/jbc.272.2.929. [DOI] [PubMed] [Google Scholar]
- Karlsson OP, Dahlqvist A, Wieslander Å. Activation of the membrane glucolipid synthesis in Acholeplasma laidlawii by phosphatidylglycerol and other anionic lipids. J Biol Chem. 1994;269:23484–23490. [PubMed] [Google Scholar]
- Kaur D, Guerin ME, Skovierova H, Brennan PJ, Jackson M. Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol. 2009;69:23–78. doi: 10.1016/S0065-2164(09)69002-X. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Dörmann P. Green light for galactolipid trafficking. Curr Opin Plant Biol. 2004;7:262–269. doi: 10.1016/j.pbi.2004.03.009. doi:10.1016/j.pbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Kim DU, Yoo JH, Lee YJ, Kim KS, Cho HS. Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar. BMB Rep. 2008;41:48–54. doi: 10.5483/bmbrep.2008.41.1.048. doi:10.5483/BMBRep.2008.41.1.048. [DOI] [PubMed] [Google Scholar]
- Klocek G, Schulthess T, Shai Y, Seelig J. Thermodynamics of melittin binding to lipid bilayers. Aggregation and pore formation. Biochemistry. 2009;48:2586–2596. doi: 10.1021/bi802127h. doi:10.1021/bi802127h. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Narise T, Sonoike K, Hashimoto H, Sato N, Kondo M, Nishimura M, Sato M, Toyooka K, Sugimoto K, et al. Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 2013;73:250–261. doi: 10.1111/tpj.12028. doi:10.1111/tpj.12028. [DOI] [PubMed] [Google Scholar]
- Korduláková J, Gilleron M, Mikusova K, Puzo G, Brennan PJ, Gicquel B, Jackson M. Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J Biol Chem. 2002;277:31335–31344. doi: 10.1074/jbc.M204060200. doi:10.1074/jbc.M204060200. [DOI] [PubMed] [Google Scholar]
- Kowarik M, Numao S, Feldman MF, Schulz BL, Callewaert N, Kiermaier E, Catrein I, Aebi M. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 2006;314:1148–1150. doi: 10.1126/science.1134351. doi:10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- Kozlov MM. Biophysics: Joint effort bends membrane. Nature. 2010;463:439–440. doi: 10.1038/463439a. doi:10.1038/463439a. [DOI] [PubMed] [Google Scholar]
- Kozmon S, Tvaroska I. Catalytic mechanism of glycosyltransferases: Hybrid quantum mechanical/molecular mechanical study of inverting N-acetylglucosaminyltransferase I. J Am Chem Soc. 2006;128:16921–16927. doi: 10.1021/ja065944o. doi:10.1021/ja065944o. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions and mechanisms. Ann Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. Review doi:10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Larivière L, Moréra S. Structural evidence of a passive base-flipping mechanism for beta-glucosyltransferase. J Biol Chem. 2004;279:34715–34720. doi: 10.1074/jbc.M404394200. doi:10.1074/jbc.M404394200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee BI, Suh SW. Crystal structure of the catalytic domain of cholesterol-α-glucosyltransferase from Helicobacter pylori. Proteins. 2011;79:2321–2326. doi: 10.1002/prot.23038. doi:10.1002/prot.23038. [DOI] [PubMed] [Google Scholar]
- Lee SS, Hong SY, Errey JC, Izumi A, Davies GJ, Davis BG. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat Chem Biol. 2011;7:631–638. doi: 10.1038/nchembio.628. doi:10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. doi:10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lesk AM. NAD-binding domains of dehydrogenases. Curr Opin Struct Biol. 1995;5:775–783. doi: 10.1016/0959-440x(95)80010-7. doi:10.1016/0959-440X(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Leventis R, Silvius JR. Lipid-binding characteristics of the polybasic carboxy-terminal sequence of K-ras4B. Biochemistry. 1998;37:7640–7648. doi: 10.1021/bi973077h. doi:10.1021/bi973077h. [DOI] [PubMed] [Google Scholar]
- Li L, Storm P, Karlsson OP, Berg S, Wieslander Å. Irreversible binding and activity control of the 1, 2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii at an anionic lipid bilayer surface. Biochemistry. 2003;42:9677–9686. doi: 10.1021/bi034360l. doi:10.1021/bi034360l. [DOI] [PubMed] [Google Scholar]
- Lind J, Ramo T, Klement R, Barany-Wallje E, Epand RM, Epand RF, Mäler L, Wieslander Å. High cationic charge and bilayer interface-binding helices in a regulatory lipid glycosyltransferase. Biochemistry. 2007;46:5664–5677. doi: 10.1021/bi700042x. doi:10.1021/bi700042x. [DOI] [PubMed] [Google Scholar]
- Lindblom G, Brentel I, Sjölund M, Wikander G, Wieslander Å. Phase equilibria of membrane lipids from Acholeplasma laidlawii: Importance of a single lipid forming nonlamellar phases. Biochemistry. 1986;25:7502–7510. doi: 10.1021/bi00371a037. doi:10.1021/bi00371a037. [DOI] [PubMed] [Google Scholar]
- Lindblom G, Hauksson JB, Rilfors L, Bergenståhl B, Wieslander Å, Eriksson PO. Membrane lipid regulation in Acholeplasma laidlawii grown with saturated fatty acids. Biosynthesis of a triacylglucolipid forming reversed micelles. J Biol Chem. 1993;268:16198–16207. [PubMed] [Google Scholar]
- Lin LY, Rakic B, Chiu CP, Lameignere E, Wakarchuk WW, Withers SG, Strynadka NC. Structure and mechanism of the lipooligosaccharide sialyltransferase from Neisseria meningitidis. J Biol Chem. 2011;286:37237–37248. doi: 10.1074/jbc.M111.249920. doi:10.1074/jbc.M111.249920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Haase AM, Lindqvist L, Lindberg AA, Reeves PR. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: Identification and characterization of transferase genes of groups B, C2, and E1. J Bacteriol. 1993;175:3408–3413. doi: 10.1128/jb.175.11.3408-3413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. doi:10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–355. doi: 10.1038/nature10151. doi:10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- Lokappa SB, Ulmer TS. Alpha-synuclein populates both elongated and broken helix states on small unilamellar vesicles. J Biol Chem. 2011;286:21450–7. doi: 10.1074/jbc.M111.224055. doi:10.1074/jbc.M111.224055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. doi:10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- Lovering AL, Lin LY, Sewell EW, Spreter T, Brown ED, Strynadka NC. Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis. Nat Struct Mol Biol. 2010;17:582–589. doi: 10.1038/nsmb.1819. doi:10.1038/nsmb.1819. [DOI] [PubMed] [Google Scholar]
- Lu J, Takahashi T, Ohoka A, Nakajima K, Hashimoto R, Miura N, Tachikawa H, Gao XD. Alg14 organizes the formation of a multiglycosyltransferase complex involved in initiation of lipid-linked oligosaccharide biosynthesis. Glycobiology. 2012;22:504–516. doi: 10.1093/glycob/cwr162. doi:10.1093/glycob/cwr162. [DOI] [PubMed] [Google Scholar]
- Luckey M. Membrane Structural Biology: with Biochemical and Biophysical Foundations. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Martinez-Fleites C, Proctor M, Roberts S, Bolam DN, Gilbert HJ, Davies GJ. Insights into the synthesis of lipopolysaccaride and antibiotics through the structures of two retaining glycosyltransferases from family GT4. Chem Biol. 2006;13:1143–1152. doi: 10.1016/j.chembiol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. The electrostatic properties of membranes. Annu Rev Biophys Biophys Chem. 1989;18:113–136. doi: 10.1146/annurev.bb.18.060189.000553. Review doi:10.1146/annurev.bb.18.060189.000553. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. Review doi:10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. doi:10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Meier M, Seelig J. Thermodynamics of the coil-beta-sheet transition in a membrane environment. J Mol Biol. 2007;369:277–289. doi: 10.1016/j.jmb.2007.02.082. doi:10.1016/j.jmb.2007.02.082. [DOI] [PubMed] [Google Scholar]
- Miley MJ, Zielinska AK, Keenan JE, Bratton SM, Radominska-Pandya A, Redinbo MR. Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J Mol Biol. 2007;369:498–511. doi: 10.1016/j.jmb.2007.03.066. doi:10.1016/j.jmb.2007.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. doi:10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C. Galactoglycerolipid metabolism under stress: A time for remodeling. Trends Plant Sci. 2011;16:98–107. doi: 10.1016/j.tplants.2010.11.004. Review doi:10.1016/j.tplants.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Monegal A, Planas A. Chemical rescue of α3-galactosyltransferase. Implications in the mechanism of retaining glycosyltransferases. J Am Chem Soc. 2006;128:16030–16031. doi: 10.1021/ja0659931. doi:10.1021/ja0659931. [DOI] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: Facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. doi:10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita YS, Fukuda T, Sena CB, Yamaryo-Botte Y, McConville MJ, Kinoshita T. Inositol lipid metabolism in mycobacteria: Biosynthesis and regulatory mechanisms. Biochim Biophys Acta. 2011;1810:630–641. doi: 10.1016/j.bbagen.2011.03.017. Review doi:10.1016/j.bbagen.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Morita YS, Velasquez R, Taig E, Waller RF, Patterson JH, Tull D, Williams SJ, Billman-Jacobe H, McConville MJ. Compartmentalization of lipid biosynthesis in mycobacteria. J Biol Chem. 2005;280:21645–21652. doi: 10.1074/jbc.M414181200. doi:10.1074/jbc.M414181200. [DOI] [PubMed] [Google Scholar]
- Ni L, Chokhawala HA, Cao H, Henning R, Ng L, Huang S, Yu H, Chen X, Fisher AJ. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. doi:10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
- Ni L, Sun M, Yu H, Chokhawala H, Chen X, Fisher AJ. Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry. 2006;45:2139–2148. doi: 10.1021/bi0524013. doi:10.1021/bi0524013. [DOI] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: Sweeter than ever. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. doi:10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Oriol R, Martinez-Duncker I, Chantret I, Mollicone R, Codogno P. Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol Biol Evol. 2002;19:1451–1463. doi: 10.1093/oxfordjournals.molbev.a004208. doi:10.1093/oxfordjournals.molbev.a004208. [DOI] [PubMed] [Google Scholar]
- Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: A complex membrane-anchoring structure for proteins. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. Review doi:10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson K, Ly HD, Dieckelmann M, Wakarchuk WW, Withers SG, Strynadka NC. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol. 2001;8:166–175. doi: 10.1038/84168. doi:10.1038/84168. [DOI] [PubMed] [Google Scholar]
- Quiros LM, Carbajo BJ, Brana AF, Salas JA. Glycosylation of macrolide antibiotics: Purification and kinetic studies of a macrolide glycosyltransferase from Streptomyces antibioticus. J Biol Chem. 2000;275:11713–11720. doi: 10.1074/jbc.275.16.11713. doi:10.1074/jbc.275.16.11713. [DOI] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Ann Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. doi:10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadurai S, Holt A, Krasnikov V, van den Bogaart G, Killian JA, Poolman B. Lateral diffusion of membrane proteins. J Am Chem Soc. 2009;131:12650–6. doi: 10.1021/ja902853g. doi:10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Boeggeman E, Ramasamy V, Qasba PK. Structure and catalytic cycle of β-1,4-galactosyltransferase. Curr Opin Struct Biol. 2004;14:593–600. doi: 10.1016/j.sbi.2004.09.006. doi:10.1016/j.sbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Rojas-Cervellera V, Ardèvol A, Boero M, Planas A, Rovira C. Formation of a covalent glycosyl-enzyme species in a retaining glycosyltransferase. Chem A Eur J. 2013;19:14018–14023. doi: 10.1002/chem.201302898. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding proteins. Nature. 1974;250:194–195. doi: 10.1038/250194a0. doi:10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Hansen G, Singh S, Hanuszkiewicz A, Lindner B, Fukase K, Woodard RW, Holst O, Hilgenfeld R, Mamat U, et al. Structural and mechanistic analysis of the membrane-embedded glycosyltransferase WaaA required for lipopolysaccharide synthesis. Proc Natl Acad Sci USA. 2012;109:6253–6258. doi: 10.1073/pnas.1119894109. doi:10.1073/pnas.1119894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm VL. Enzymatic Transition States, transition-state analogs, dynamics, thermodynamics, and lifetimes. Annu Rev Biochem. 2011;80:703–732. doi: 10.1146/annurev-biochem-061809-100742. doi:10.1146/annurev-biochem-061809-100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. Thermodynamics of lipid-peptide interactions. Biochim Biophys Acta. 2004;1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. Review doi:10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Sheng F, Jia X, Yep A, Preiss J, Geiger JH. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J Biol Chem. 2009;284:17796–17807. doi: 10.1074/jbc.M809804200. doi:10.1074/jbc.M809804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soya N, Fang Y, Palcic MM, Klassen JS. Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology. 2011;21:547–552. doi: 10.1093/glycob/cwq190. doi:10.1093/glycob/cwq190. [DOI] [PubMed] [Google Scholar]
- Spiro RG. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. doi:10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- Steiner K, Hagelueken G, Messner P, Schaeffer C, Naismith JH. Structural basis of substrate binding in WsaF, a rhamnosyltransferase from Geobacillus stearothermophilus. J Mol Biol. 2010;397:436–447. doi: 10.1016/j.jmb.2010.01.035. doi:10.1016/j.jmb.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Palaniappan A, Duncan K, Rhoads DM, Huber SC, Sachs MM. Mitochondrial localization and putative signaling function of sucrose synthase in maize. J Biol Chem. 2006;281:15625–15635. doi: 10.1074/jbc.M600355200. doi:10.1074/jbc.M600355200. [DOI] [PubMed] [Google Scholar]
- Sun HY, Lin SW, Ko TP, Pan JF, Liu CL, Lin CN, Wang AH, Lin CH. Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design. J Biol Chem. 2007;282:9973–9982. doi: 10.1074/jbc.M610285200. doi:10.1074/jbc.M610285200. [DOI] [PubMed] [Google Scholar]
- Szpryngiel S, Ge C, Iakovleva I, Georgiev A, Lind J, Wieslander Å, Mäler L. Lipid interacting regions in phosphate stress glycosyltransferase atDGD2 from Arabidopsis thaliana. Biochemistry. 2011;50:4451–4466. doi: 10.1021/bi200162f. doi:10.1021/bi200162f. [DOI] [PubMed] [Google Scholar]
- Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. doi:10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. Review doi:10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Takakura Y, Yamamoto T. Purification, cloning, and expression of an alpha/beta-galactoside alpha-2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. J Biol Chem. 2007;282:29794–29802. doi: 10.1074/jbc.M701907200. doi:10.1074/jbc.M701907200. [DOI] [PubMed] [Google Scholar]
- Urresti S, Albesa-Jové D, Schaeffer F, Pham HT, Kaur D, Gest P, van der Woerd MJ, Carreras-González A, López-Fernández S, Alzari PM, et al. Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria. J Biol Chem. 2012;287:24649–24661. doi: 10.1074/jbc.M112.368191. doi:10.1074/jbc.M112.368191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi CR, Hart G, Etzler ME. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- Vetting MW, Frantom PA, Blanchard JS. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: Substrate-assisted catalysis. J Biol Chem. 2008;283:15834–15844. doi: 10.1074/jbc.M801017200. doi:10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom S, Li L, Karlsson OP, Wieslander Å. Key role of the diglucosyldiacylglycerol synthase for the nonbilayer-bilayer lipid balance of Acholeplasma laidlawii membranes. Biochemistry. 1999;38:5511–5520. doi: 10.1021/bi982532m. doi:10.1021/bi982532m. [DOI] [PubMed] [Google Scholar]
- Vrielink A, Ruger W, Driessen HP, Freemont PS. Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13:3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Weldeghiorghis T, Zhang G, Imperiali B, Prestegard JH. Solution structure of Alg13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Structure. 2008;16:965–975. doi: 10.1016/j.str.2008.03.010. doi:10.1016/j.str.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–287. doi: 10.1038/nrmicro1861. doi:10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. doi:10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Wieslander Å, Rilfors L, Lindblom G. Metabolic changes of membrane lipid composition in Acholeplasma laidlawii by hydrocarbons, alcohols, and detergents: Arguments for effects on lipid packing. Biochemistry. 1986;25:7511–7517. doi: 10.1021/bi00371a038. doi:10.1021/bi00371a038. [DOI] [PubMed] [Google Scholar]
- Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. Review doi:10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- Wrabl JO, Grishin NV. Homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily. J Mol Biol. 2001;314:365–374. doi: 10.1006/jmbi.2001.5151. doi:10.1006/jmbi.2001.5151. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proc Natl Acad Sci USA. 2007;104:5348–5353. doi: 10.1073/pnas.0701160104. doi:10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Anderson S, Zhang Y, Garavito RM. The structure of sucrose synthase-1 from Arabidopsis thaliana and its functional implications. J Biol Chem. 2011;286:36108–36118. doi: 10.1074/jbc.M111.275974. doi:10.1074/jbc.M111.275974. [DOI] [PMC free article] [PubMed] [Google Scholar]