Figure 1.
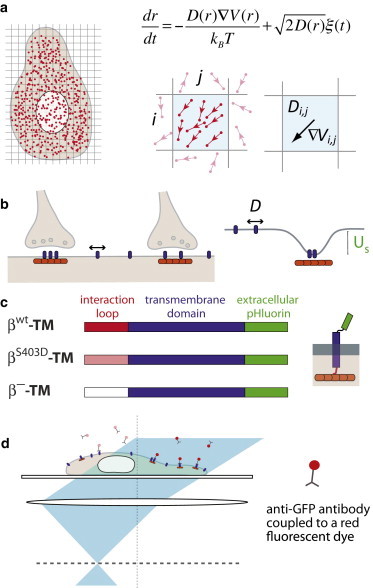
General scheme of the assay. (a) Principle of the Bayesian inference method. (Left) High-density single-molecule data (red dots) are recorded at the cell surface. (Right) In a mesh domain, multiple translocations (top) are used to infer the local diffusivity and force (gradient of the potential) that underlie the motion (bottom). (b) GlyRs (blue) diffuse in the membrane and are in dynamic equilibrium between synaptic and extrasynaptic domains in the neuronal membrane. At synapses, GlyRs are stabilized by their interactions with gephyrin clusters (orange), which can be modeled as trapping potential (with depth US). (c) Expression constructs of transmembrane proteins with an extracellular pHluorin tag and an intracellular interaction loop derived from the GlyR β-subunit. (d) Principle of high-density single-molecule uPAINT imaging (16). To see this figure in color, go online.
