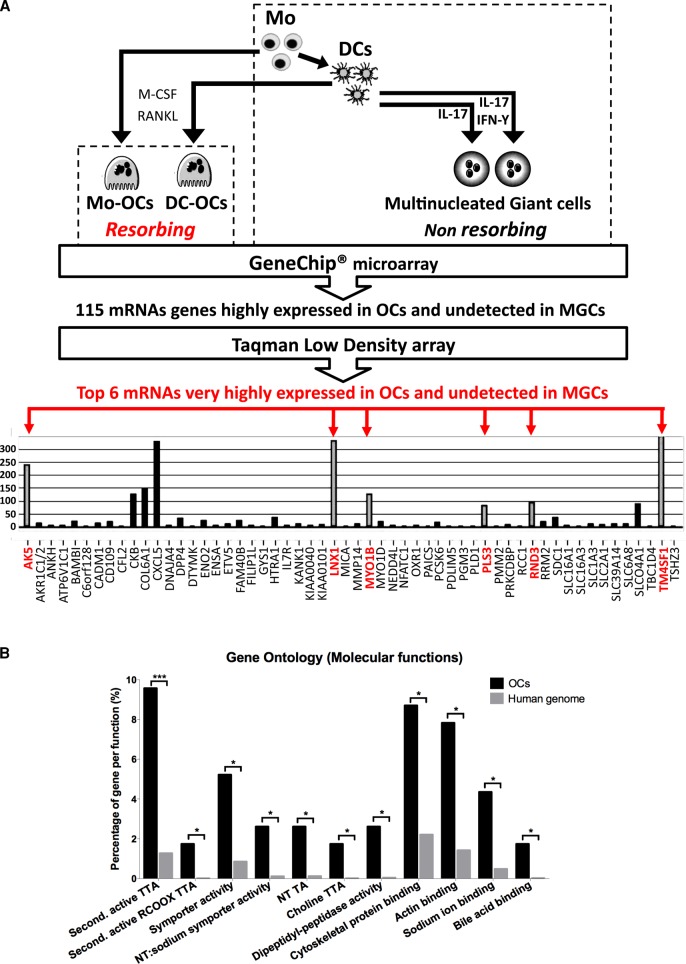
FIGURE 3:
Workflow and results of the comparative transcriptomic study used to probe for OC-specific genes. (A) In vitro treatments used for differentiation and fusion of human Mos into resorbing OCs, of immature DCs into resorbing OCs, and of DCs into nonresorbing MGCs by addition of IL-17 and/or IFN-γ. GeneChip Microarray technique was applied to total RNAs obtained from each of cell type, followed by mathematical comparison to sort out 115 mRNAs highly and exclusively expressed in Mo-OCs and DC-OCs. Validation of the up-regulation of the 115 genes in OCs by Taqman low-density array performed on cDNAs from Mos, DCs, Mo-OCs, DC-OCs, and DC-17γ-MGCs. Due to the high cutoff value for detection, only 56 of 115 genes were found to be expressed in OCs. Among these, six genes (gray columns, names in red) were undetected in Mos, DCs, or in MGCs. (B) Enriched Gene Ontology molecular functions in the 115 OC-specific genes compared with the human genome identified by the online tool FatiGO. NT, neurotransmitter; second, secondary; TA, transporter activity; TTA, transmembrane transporter activity.