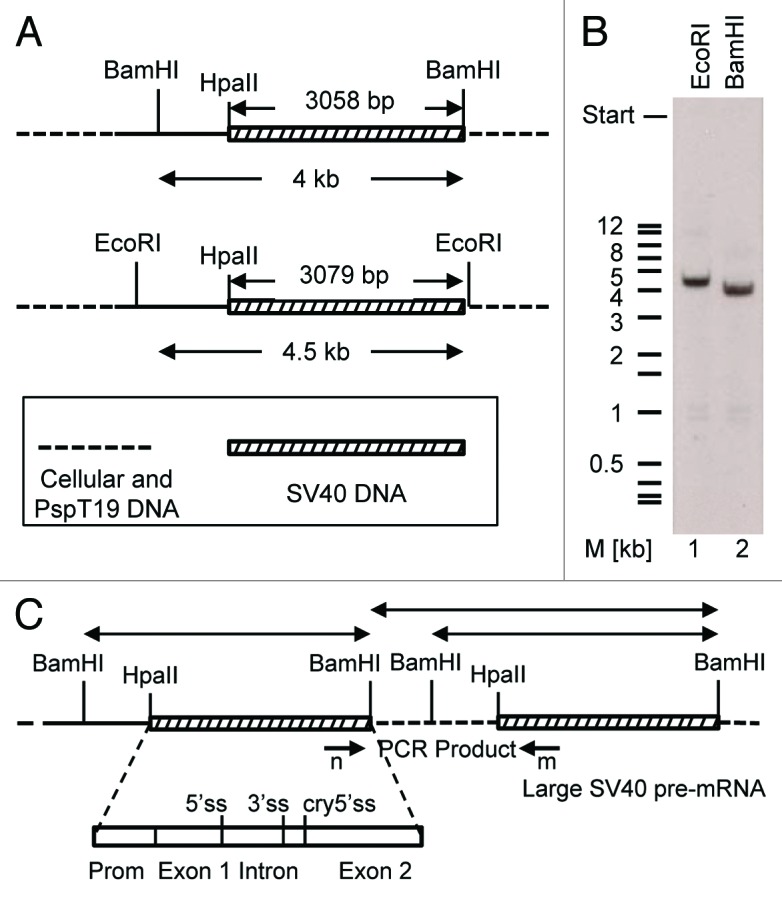
Figure 5. Sequence duplication in the sT-ag-associated transcript was not related to an equivalent DNA duplication. (A) Enzymatic cleavage scheme of genomic DNA for southern hybridization. The genomic DNA of a sT-ag producing clone was digested with EcoRI or BamHI. Both enzymes cut outside the SV40 DNA coding for T-ag: once in the multiple cloning site of the vector pspT19 that was used for cloning of the SV40 HpaII–BamHI early SV40 DNA segment reconstituting the BamHI site, and once upstream in a distance of about 3.1 kb (size of the HpaII-BamHI fragment) or more within the host genomic DNA. (B) Single bands of about 4.5 kb (EcoRI, lane 1) or 4 kb (BamHI, lane 2) indicated the integration of a single copy of the early SV40 DNA HpaII–BamHI segment into the genomic DNA of the host cell. (C) Hypothetical HpaII–BamHI SV40 DNA segment tandem duplication. BamHI (or EcoRI) digestion of the genomic DNA would generate two SV40-specific BamHI fragments. If the host-genomic DNA linker sequence does not contain a BamHI site, then the size of one fragment would indirectly indicate the distance of the two integrated HpaII–BamHI SV40 DNA segments in the genomic host DNA. A PCR from the genomic DNA with primers n and m, binding to the 3′end of exon 1 and the 5′end of exon 2, respectively, using conditions which favor the amplification of long DNA fragments, did not give any product. These results exclude a tandem integration of SV40 DNA into the genome of the sT-ag-producing host cell clone.
