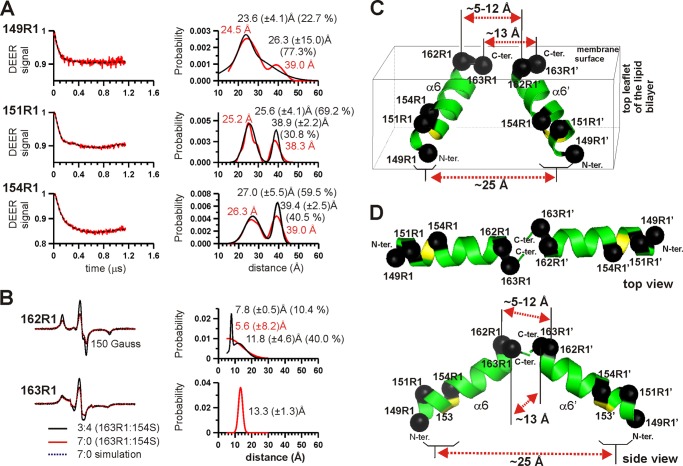
FIGURE 6.
Proximity of helix α6 residues in membrane-inserted BAK oligomers. A, DEER data for residues 149R1, 151R1, and 154R1 in the absence of spin dilution. DEER experiments were carried out for the indicated proteins without spin dilution. The DEER data (left column) were analyzed by the DeerAnalysis program using Tikhonov regularization, resulting in the distance distribution functions on the right panel (red traces). The black dotted lines superimposed to the DEER signals (red traces, left panel) are the calculated DEER signal for the distance fit. Results by two Gaussian models are superimposed (black traces, right panel) with the average inter-spin distance, the standard deviation and the percent of the spin pairs for the distances indicated. B, proximity of helix α6 residues 162R1 and 163R1 in BAK oligomers. The area-normalized EPR spectra of the spin-diluted samples (3:4 mixture, black trace) of the indicated membrane-inserted BAK are superimposed to those of the undiluted samples (7:0 mixture, red trace). The spectra for 162R1 and 163R1 were acquired at −30 and 22 °C, respectively. The data could be best-fitted with one or two Gaussian distance distributions (right panel). Residue 162R1, with a spin labeling efficiency of 74.2 (±0.6)%, had the interacting spin pairs close to the expected value of 55% (52.3% interacting spins for the single Gaussian fit (red trace) and 50.4% (=10.4 + 40.0) for the double Gaussian fit (black trace). Similarly, residue 163R1 (58.6 (±1.1)% labeling efficiency), resulted in ∼35% interacting spin pairs (34% expected) in the data fitting. C and D, pseudo parallel or pseudo anti-parallel arrangement of two neighboring α6 helices in the BAK oligomer. Two tilted α6 helices are arranged side by side (C) or in a head-to-head orientation (D) with their C termini in close proximity. The average inter-spin distances (dotted arrows) are shown for the indicated pairs of residues (Cα carbon atoms in black spheres).