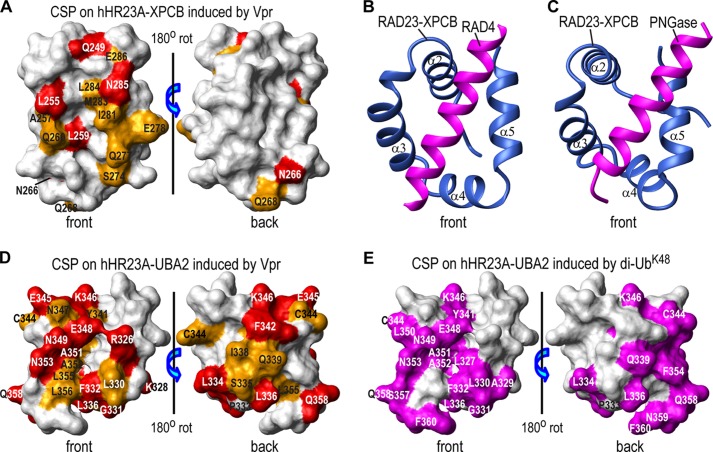
FIGURE 4.
Structural mapping of Vpr binding sites on the XPCB and UBA2 domains. Vpr binding sites are mapped onto the NMR structures of the hHR23A XPCB domain (PDB code 1TP4) (52) (A) and the hHR23A UBA2 domain (PDB code 1QZE) (51) (D). Residues that exhibit chemical shift changes of ≥0.10 ppm and between 0.05 and 0.10 ppm are colored red and orange, respectively. They are labeled with their residue names and numbers. The RAD23 XPCB domain interactions with RAD4 and PNGase, as observed in the x-ray structures of the yeast RAD23-RAD4 complex (PDB code 2QSF) (53) (B) and the RAD23-PNGase complex (PDB code 1X3W) (C), respectively. Only the XPCB domain of RAD23 (blue) and the N-terminal helices (magenta) of RAD4 (residues 141–161, B) or of PNGase (residues 10–30, C) are shown. All XPCB domains are depicted in the same orientation. E, di-UbK48 binding site mapping onto the NMR structure of the UBA2 domain of hHR23A (PDB code 1QZE) (51) with UBA2 residues that exhibit large chemical shift changes (≥0.10 ppm), colored in magenta (from Fig. 2B in Ref. 56). CSP, chemical shift perturbations; rot, rotation.