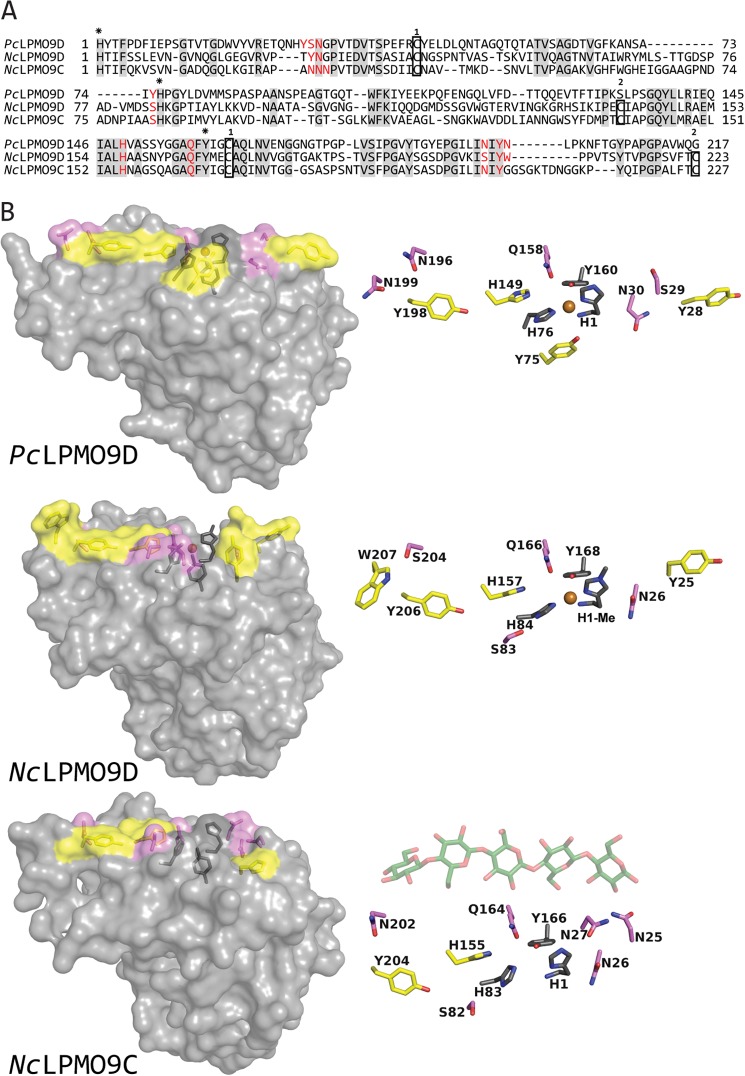
FIGURE 8.
Sequence and structural comparisons of PcLPMO9D, NcLPMO9D and NcLPMO9C. A, structure-guided sequence alignment of PcLPMO9D, NcLPMO9D, and NcLPMO9C with fully conserved residues shaded in gray. Residues involved in coordination of the copper are marked with asterisks above the sequence (His-1, His-83 and Tyr-166 in NcLPMO9C), whereas aromatic surface residues and protruding polar surface residues potentially involved in substrate binding are colored in red (all highlighted in B). Cysteines forming disulfide bonds are indicated in boxes with numbers above showing which cysteines are connected. B, side chains on the substrate binding surface of PcLPMO9D (Protein Data Bank code 4B5Q) and NcLPMO9D (Protein Data Bank code 4EIR) compared with the modeled structure of NcLPMO9C. Copper coordinating residues are colored dark gray, aromatic and protruding polar residues putatively involved in substrate binding are colored yellow and violet, respectively, and the copper is shown as an orange sphere. For illustration purposes only, a cellopentaose (coordinates derived from Protein Data Bank code 2EEX; shown in green with oxygens in red) is placed above the surface of the modeled structure of NcLPMO9C. The view in the right panels is rotated 90° relative to the view in the left panels, looking down at the flat surface containing the copper binding site.