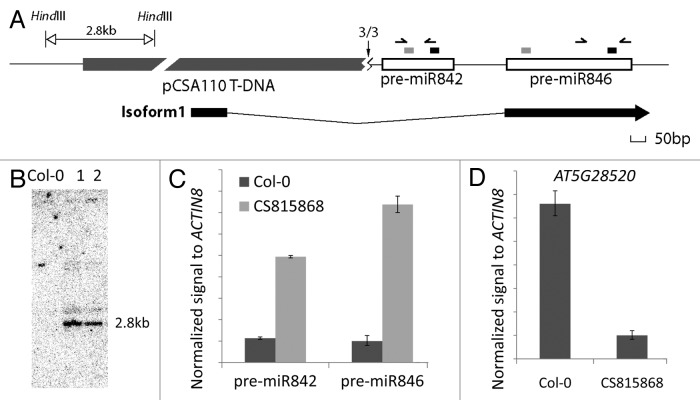
Figure 2.AT5G28520 is downregulated in CS815868 T-DNA insertion plants, which overexpress pre-miR842 and pre-miR846. (A) Genomic structure around MIR842 and MIR846 in T-DNA insertion line CS815868. TSS revealed by 5′ RACE is shown with a vertical black arrow (numbers are sequenced clones mapping to site out of the total clones sequenced). The zigzag below the arrow represents a 40 bp sequence found that does not map to the T-DNA and has homology to rDNA. Open boxes represent pre-miR842 and pre-miR846. The short gray rectangles above the open boxes represent miRNA* and short black rectangles represent mature miRNA sequences. Isoform 1, which is the most abundant splice variant expressed from this locus in wild type is shown below. Black boxes represent exons. Horizontal arrows represent primers used for real-time PCR of pre-miRNAs in (C). (B) Genomic Southern blot showing single insert in CS815868. Lanes 1 and 2 are HindIII-digested genomic DNAs from two individual CS815868 plants. Blot was probed with BASTA selectable marker gene fragment from the vector. (C and D) Real-time PCR results showing the different expression levels of AT5G28520 (C) and pre-miR842 and -miR846 (D) in roots of control Col-0 and CS815868 line. Error bar, ± s.d., n = 2.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.