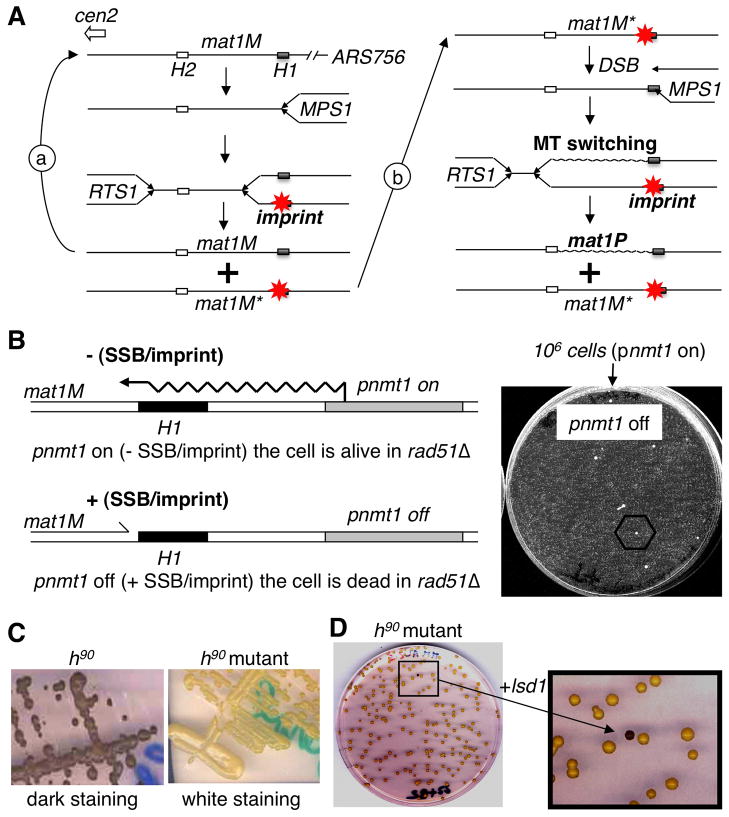
Figure 1. Selection and complementation of a new imprinting mutant.
(A) Schematic representation of two consecutive rounds of replication at mat1. Starting from a virgin mat1M locus (left). The close proximity of ARS756 with RTS1, constrains the polarity of mat1 replication to ensure optimal imprinting (*) at MPS1. Following DNA replication only one of the two mat1M loci is imprinted. During the following DNA replication: (a) replication of the virgin mat1M, (b) replication of the imprinted mat1M; the fork transforms the imprint or single-strand break into a polar one ended double-strand break (DSB) that promotes MT switching (mat1P) and the imprint is formed on the unswitched mat1M sister locus. The transformation of the imprint into a DSB requires homologous recombination (i.e. Rhp51) for cell survival.
(B) Selection of spontaneous survivors in the selection system. Left panel: By introducing the thiamine repressible promoter (pnmt1) upstream of mat1, we can force transcription through the imprinted region, erasing the imprint (-SSB), in a reversible manner (+SSB). Right panel: 106 cells grown under permissive conditions (pnmt1 on) are plated in the presence of thiamine (pnmt1 off) and only a few spontaneous mutants can form colonies.
(C) Iodine staining of mutants. By crossing the initial nbt-18 mutants with the wild-type h90 strain we introduced the mutation in the wild-type background and assayed by iodine staining. The left panel shows the dark staining of the wild-type h90 strain and the right panel shows the white staining of the h90strain carrying the mutant allele.
(D) Functional complementation. The h90 strain carrying the mutation (white) is transformed with a genomic library and colonies are assayed for coloration by iodine vapors in order to reveal the complemented cells (black colony in the enlarged view).