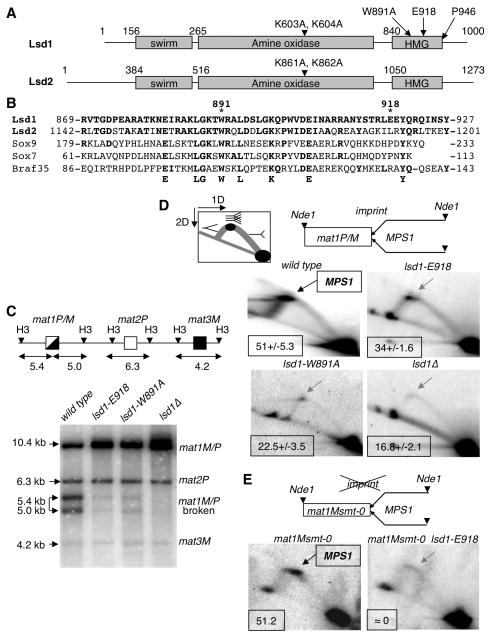
Figure 2. The Lsd1 HMG domain is required for DSB and MPS1 activity at mat1.
(A) Schematic representation of Lsd1 and Lsd2. The protein domains and the mutations in lsd1 and lsd2 are indicated.
(B) Alignment of the Lsd1 HMG domain with HMG domains from Lsd2, Sox9, Sox7 and Braf35 from mouse. The conserved residues are indicated.
(C) lsd1 mutants affect the level of the break at mat1. Top panel: Schematic representation of the mat region, showing mat1P/M, mat2P and mat3M cassettes. The sizes of the HindIII fragments (H3) are indicated (Kbp). Lower panel, Southern blot of HindIII digested DNA from the lsd1, lsd1-E918, lsd1-W891A and lsd1Δ strains, hybridized with the labeled mat1P HindIII-containing fragment. The mat1P probe also hybridizes with the mat2P and mat3M cassettes. The sizes and identities of the DNA fragments are indicated.
(D) Lsd1 is required for MPS1 activity at mat1P/M. Top panel: diagram of the migration pattern of the replication intermediates detected in 2D gel electrophoresis. The position of the imprint, MPS1 pause site and the polarity of DNA replication are indicated. DNA replication intermediates at mat1 from the wild type, lsd1-E918, lsd1-W891A and lsd1Δ strains are analyzed (lower panel). In the wild type panel the DNA replication intermediates accumulating at MPS1 are indicated by an arrow. The percentage of pause is shown for each strain from 2 to 3 independent 2D gels.
(E) Lsd1 is required for MPS1 activity in mat1Msmt-0 (deletion of 263 bp distal to mat1). DNA replication intermediates from the Msmt-0 and Msmt-0 lsd1-E918 strains were analyzed and show the decrease of MPS1 activity in the lsd1-E918 background.