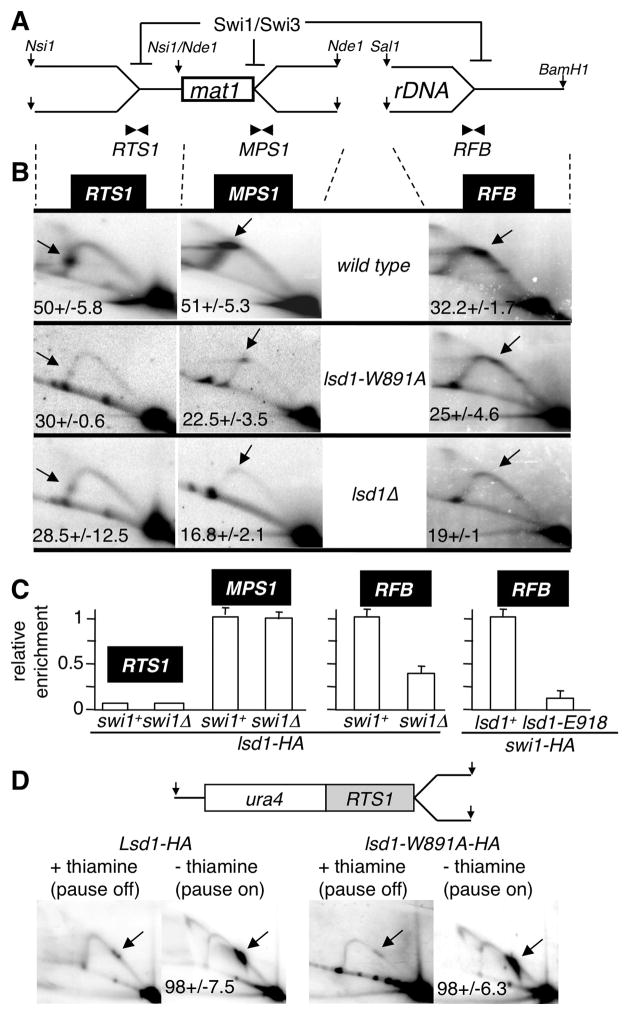
Figure 5. Lsd1 controls directly RFB and indirectly RTS1 replication fork barriers.
(A) Schematic representation of the Swi1/Swi3-dependent DNA replication fork pauses and arrests on the proximal (RTS1) and distal (MPS1) sides of mat1 and at the rDNA (RFB) loci. The restriction enzymes used for genomic digestions are indicated.
(B) 2D gel autoradiograms of RTS1 (left), MPS1 (middle) and RFB (right) in the wild type (top), lsd1-W891A (middle) and lsd1Δ (bottom) strains. The restriction enzymes used are indicated. Arrows show the accumulating DNA replication material at pause/arrest sites.
(C) ChIP analysis of Lsd1-HA at RTS1, MPS1 and RFB. Quantitative PCR analysis of the ChIP of lsd1-HA in swi1+ and swi1Δ backgrounds using primer pairs close to the pause/arrest sites (RTS1, MPS1 and RFB). On the left panel, Lsd1-HA swi1+ was set to 1 at MPS1 (as a single copy locus) and on the right panel, Lsd1-HA swi1+ was set to 1 at RFB, independently due to the high rDNA copy number.
(D) RTS1 activity is lsd1-independent. Schematic representation of the Ase1 restriction fragment containing the RTS1 pause at the ura4 locus on chromosome 3 and 2D gel autoradiograms. RTS1 activity is “off” by repressing (+ thiamine) and “on” by inducing (- thiamine) expression of the barrier protein Rtf1, as indicated. 2D gels were probed using the ura4 fragment.