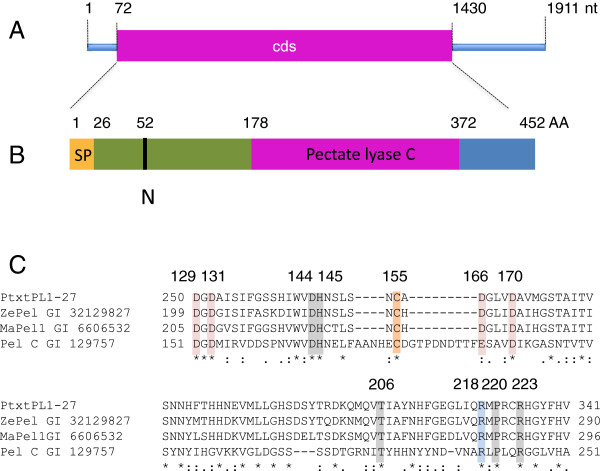
Figure 1.
Structural features of PtxtPL1-27. (A) Structure of mRNA [GenBank: EU379971.1]. Cds, coding sequence; nt, nucleotides. (B) Structure of the deduced peptide showing the signal peptide (SP), the pectate lyase C catalytic domain, and the predicted N-glycosylation site (N). AA, amino acids. (C) A multiple alignment of the catalytic site of PtxtPL1-27 and pectate lyases of known enzymatic activities, showing the conserved residues involved in Ca+2 binding (pink), disulfide bonds (orange), catalysis (blue), and substrate binding (gray) with numbering according to Pel C structure as in [16]. ZePel, Zinnia elegans pectate lyase; MaPel1, Musa acuminata pectate lyase 1; Pel C, Erwinia chrysanthemi pectate lyase C.