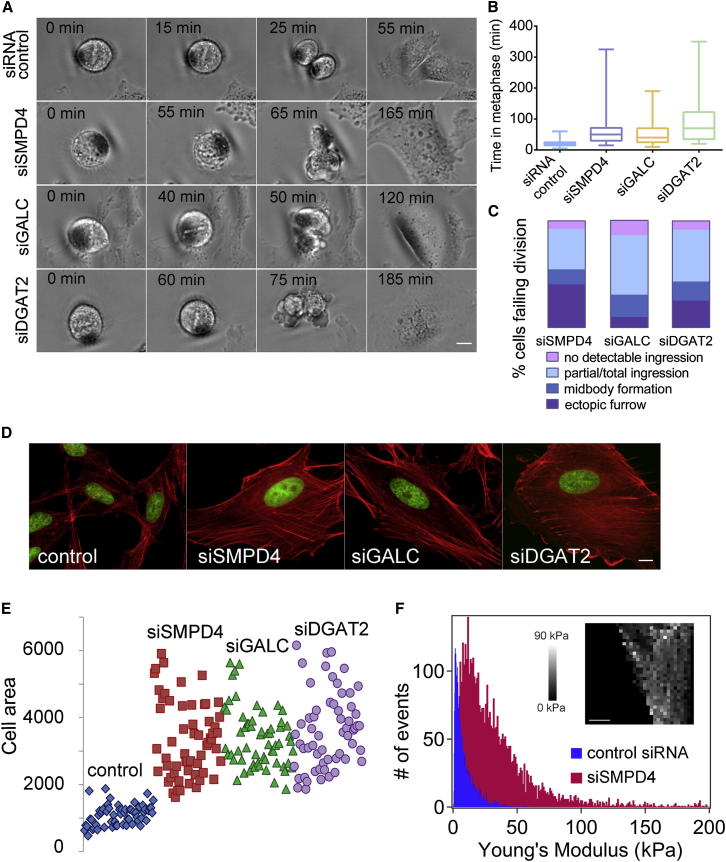
Figure 3.
RNAi Knockdown of Lipid Biosynthetic Enzymes Causes Cell Division Defects and Cytoskeletal Changes in Interphase Cells
(A) Still images from time-lapse movies of dividing cells for control, SMPD4, GALC, and DGAT2 siRNA treatments. Movies are available in the Supplemental Information. Scale bar, 10 μm.
(B) Box plots of metaphase delays caused by siSMPD4, siGALC, and siDGAT2 treatments, respectively (average ± SEM), 58 ± 4.6, 56.5 ± 5.4, and 90.6 ± 9 min. The time in metaphase for all RNAi-treated cells was significantly higher than the control (20.4 ± 1 min) with a > 99.99% confidence (t test). A minimum of 70 cells from 3 independents experiments for each case were analyzed.
(C) SMPD4, GALC, and DGAT2 siRNA treatments result in different cytokinesis failure phenotypes. Examples for each phenotype are shown in Figure S3D. Ectopic furrow refers to both successful and unsuccessful abscissions. A minimum of 70 cells from three independent experiments for each case were analyzed.
(D–F) SMPD4, GALC, and DGAT2 siRNA treatments result in altered cell shape and actin morphology.
(D) Representative images are shown for control and siRNA-treated cells. F-actin is shown in red (phalloidin staining) and DNA in green (DAPI staining). Scale bar, 10 μm.
(E) Quantification of the cell area for control cells (1088 ± 39 μm2, average ± SEM) and different siRNA-treated cells (respectively, for siSMPD4, siGALC, and siDGAT2: 3331 ± 143, 3386 ± 113, and 3617 ± 144 μm2). The area was measured in at least 60 mononucleated cells per treatment and was significantly higher in all RNAi-treated cells compared to control, with a >99.99% confidence (t test). Targeted lipid analyses of the RNAi samples are reported in Table S3 and Figure S3A.
(F) Histogram of cell stiffness (Young’s modulus values, see Extended Experimental Procedures) measured on control and SMPD4 siRNA-treated cells. A total number of 4,098 indentations (21 cells) and 4,314 indentations (18 cells) were performed on control siRNAi and siSMPD4-treated cells, respectively. The average value for siSMPD4 cells is significantly higher (35 ± 8.8 kPa, average ± SEM) than siRNA control cells (8.3 ± 1.9 kPa), with a >99.95% confidence. (Inset) Typical 32 × 32 force-volume map measured on a siSMPD4 cell. The topography of the force-volume corresponds to an arbitrary color scale of the Young’s Modulus. Scale bar, 10 μm.