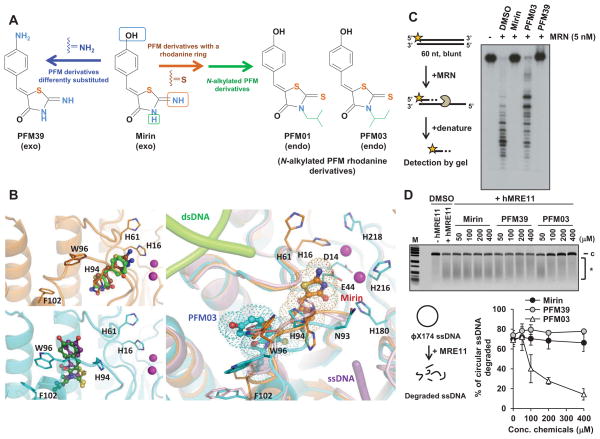
Figure 1. Development of Inhibitors against MRE11 Exo- or Endonuclease Activity.
(A) Derivation by modification of mirin. (B) TmMre11 structures in complex with exo- and endo-nuclease inhibitors. The top left panel shows an overlay of co-crystal structures of TmMre11 (cartoon, coloured brown) in complex with the exonuclease inhibitors, mirin (sticks, carbon atoms coloured brown) and PFM39 (green). The bottom left panel shows an overlay of the co-crystal structures of TmMre11 (cyan) in complex with the endonuclease inhibitors PFM01 (purple) and PFM03 (dark green). Each inhibitor type has a distinct binding site, in proximity to the active site metal ions. The right hand panel shows an overlay of the unliganded TmMre11 (cartoon, coloured pink) with mirin (brown) and PFM03 (cyan). The relative position of each inhibitor is highlighted by a dotted representation of its van der Waals surface. The endo/exo inhibitor binding sites are adjacent, but respectively positioned to distinctly disrupt dsDNA (light green tubes) end-opening by His61 or the ssDNA (violet tube) pathway adjacent to the loop containing Asn93 to Phe102, leading towards the active site metal ions. Nitrogen, oxygen and sulphur atoms are coloured blue, red and yellow respectively. Active site metal ions are shown as magenta spheres.
(C) Mirin and PFM39 but not PFM03 inhibit 5 nM MRN exonuclease activity. 100 nM labeled DNA were incubated with human MRN for 30 minutes at 37ºC. (D) PFM03, but not mirin or PFM39, inhibits human MRE11 endonuclease activity. Analysis using MRN in place of MRE11 is shown in Figure S1E. φX174 circular ssDNA was incubated with purified human MRE11 and DMSO or inhibitor at the indicated concentration. The inhibition assay was established in a time/protein concentration dependent manner to cause loss of ~70% circular DNA. The figure shows the % of circular ssDNA degraded relative to the control (DMSO-treated) sample. c: circular DNA, *: degraded DNA. PFM01 was excluded from inhibitor analysis due to inefficient solubility in vitro. Error bars represent SEM from >3 experiments. Figure S1G–I shows the impact of the inhibitors on MRN DNA binding and ATPase activity. See also Figure S1.