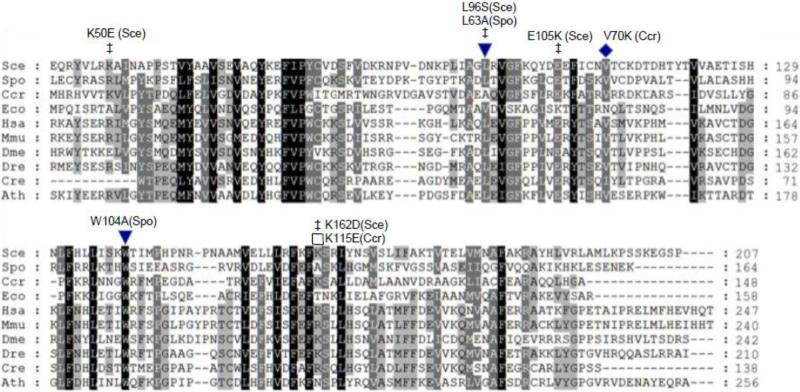
Figure 1.
Conserved amino acid residues in Coq10 polypeptide homologues. Protein sequences were aligned with BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) (Ibis Biosciences, Carlsbad, CA) and shaded as described by Genedoc with three levels of shading (http://www.nrbsc.org/gfx/genedoc/) [54]. Residues conserved in all proteins are shaded black, in 80% dark grey, and in 60% light grey. Amino-terminal segments of eukaryotic polypeptides preceding the first methionine of the C. crescentus (Ccr) and E. coli (Eco) polypeptides are not conserved and were omitted from the alignment for clarity. Residues determined to be important for maintenance of respiration are designated with filled symbols and include C. crescentus V70K (this work), S. pombe L63A and W104A [11]. Additional mutations affecting S. cerevisiae Coq10p function are designated by ‡ and include K50E, L96S, E105K, and K162D [18]. Mutation of K115E in CC1736 (marked by an open square) did not impair rescue of the S. cerevisiae coq10 null mutant (this work). The aligned sequences include: S. cerevisiae COQ10 (NCBI GeneID: 854154), Schizosaccharomyces pombe COQ10 (942096), C. crescentus CBL5 (942096), E. coli yfjG (945614), Homo sapiens COQ10A (93058), M. musculus COQ10B (67876), Drosophila melanogaster CG9410 (35568), Danio rerio Zgc:73324 (393762), Chlamydomonas reinhardtii COQ10 (5718019), and Arabidopsis thaliana AT4G17650 (827485).