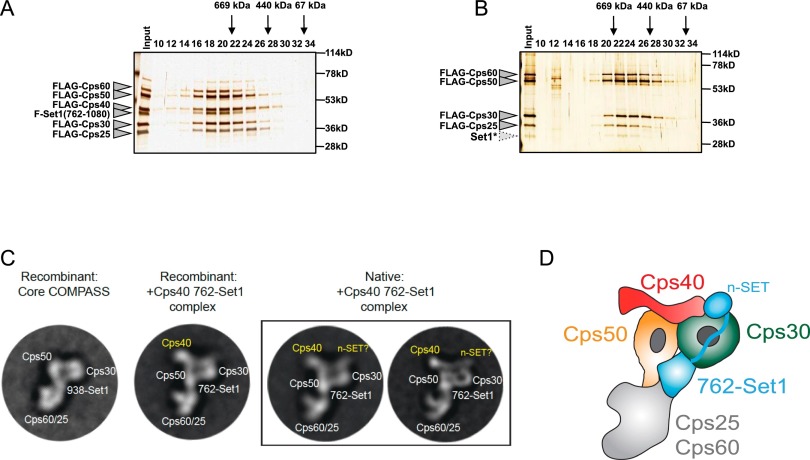
Figure 2.
Structural basis for Cps40/Spp1 stabilization of 762-Set1. (A,B). Flag-tagged 762-Set1 and other Flag-tagged COMPASS subunits were coexpressed in Sf9 cells using the baculovirus system. (A) When Flag-Cps40/Spp1 is included in the reconstitution, the assembled complex is intact, as determined by Superose 6 gel filtration chromatography, eluting at ∼800 kDa (peaking at fraction 18). (B) When Flag-Cps40/Spp1 is left out of the reconstitution, the Flag-purified complex displays an altered Superose 6 elution profile, peaking at ∼600 kDa (fraction 22). Molecular weight markers and COMPASS subunits are indicated. A truncated version of Set1 that coelutes with the COMPASS core subunits in fraction 22 is labeled as Set1* and likely corresponds to the minimal SET domain. We predict that this form of Set1 (core Set1) is generated when the n-SET domain of 762-Set1 is degraded in the absence of Cps40/Spp1. (C,D) Representative 2D class averages of COMPASS with Cps40/Spp1. (C) Representative 2D class averages of the recombinant “core” COMPASS (Cps60/Bre2, Cps50/Swd1, Cps30/Swd3, Cps25/Sdc1, and 938-Set1) are compared with class averages of recombinant “+Cps40/Spp1, 762-Set1” COMPASS (Cps60/Bre2, Cps50/Swd1, Cps40/Spp1, Cps30/Swd3, Cps25/Sdc1, and 762-Set1) and with native “+Cps40/Spp1, 762-Set1” COMPASS. Additional densities attributed to Cps40/Spp1 and n-SET are observed on top of Cps50/Swd1 and Cps30/Swd3. (D) Model of Cps60/Bre2-, Cps50/Swd1-, Cps40/Spp1-, Cps30/Swd3-, Cps25/Sdc1-, and 762-Set1-containing COMPASS with subunits and domains indicated.